
Mikrobielle Naturstoffe
Prof. Dr. Rolf Müller
Naturstoffe aus Mikroorganismen sind nach wie vor eine der vielversprechendsten Quellen für die Entwicklung neuer Wirkstoffe. Unsere Abteilung beschäftigt sich mit der Entdeckung, sowie der biotechnologischen Verbesserung und Produktion antimikrobieller Naturstoffe aus Myxobakterien.
Unsere Forschung
Myxobakterien sind dazu in der Lage zahlreiche Naturstoffe mit hoher struktureller und funktioneller Diversität zu produzieren. Bisher identifizierte myxobakterielle Naturstoffe zeigen Aktivitäten gegen eine Vielzahl an Organismen, von Bakterien und Pilzen bis hin zu Säugerzellen und Viren. Wir verwenden sowohl aktivitäts- als auch strukturgeleitete Ansätze um solche bioaktiven Stoffe zu identifizieren. Darüber hinaus benutzen wir auch moderne Methoden des genome mining, um das biosynthetische Potential wenig erforschter Myxobakterien zu erkunden. Im Anschluss an die Identifizierung arbeiten wir an der Aufklärung der Struktur, des molekularen Syntheseweges sowie der Wirkweise interessanter Naturstoffe. Diese Informationen schaffen die Grundlage für die biotechnologische und chemische Verbesserung der Substanzen. Unser Ziel ist es, Leitstrukturen zu ermitteln und weiter zu entwickeln, welche möglichst schnell in klinischen Studien zum Einsatz kommen sollen. Es ist uns gelungen, eine hauseigene Stammsammlung mit mehr als 10.000 einzigartigen myxobakteriellen Stämmen aufzubauen, welche aus Proben von der ganzen Welt isoliert wurden.
Team-Mitglieder

Prof. Dr. Rolf Müller
Gruppenleiter

Christina Decker
Assistentin

Ellen Merckel
Sekretärin

Dr. Carsten Volz
Labormanager / Wissenschaftler

Dr. Daniel Krug
Wissenschaftler

Jeenu Joy
Wissenschaftlerin

Dr. Jennifer Herrmann
Wissenschaftlerin

Dr. Judith Hoffmann
Wissenschaftlerin

Dr. Ronald Garcia
Wissenschaftler

Dr. Abdelhalim Babiker Mahmoud
Postdoc

Dr. Asfandyar Sikandar
Postdoc

Dr. Carsten Seyfert
Postdoc

Dr. Feng Xie
Postdoc

Dr. Guangzhi Dai
Postdoc

Dr. Jake Haeckl
Postdoc

Dr. Jonas Baumann
Postdoc

Dr. Jordan Espenshade
Postdoc

Dr. Joy Birkelbach
Postdoc

Dr. Kamal Tehrani
Postdoc

Dr. Katarina Cirnski
Postdoc

Dr. Markus Neuber
Postdoc

Dr. Peter Sullivan
Postdoc

Dr Qiyao Shen
Postdoc

Dr. Sari Rasheed
Postdoc

Dr. Sebastian Walesch
Postdoc

Dr. Sumire Kurosawa
Postdoc

Dr. Susanne Kirsch-Dahmen
Postdoc

Dr. Tatiana Gorelik
Postdoc

Dr. Yuhao Ren
Postdoc

Dr. Yunsheng Gao
Postdoc

Alexander Voltz
Doktorand

Alison Müller
Doktorandin

Anna Kastner
Doktorandin

Anna-Lena Huber
Doktorandin

Anna-Lisa Jäckel
Doktorandin

Christoph Porten
Doktorand

Clemens Schumm
Doktorand

Emine Erdogan Ozeker
Doktorandin

Fabienne Wittling
Doktorandin

Felix Deschner
Doktorand

Franziska Fries
Doktorandin

Jessica Lamber
Doktorandin

Kai Schließmann
Doktorand

Laxmi Neupane
Doktorandin

Mary Victory Gutierrez
Doktorandin

Márió Széles
Doktorand

Philipp Donate
Doktorand

Roman Reberšek
Doktorand

Selina Deckarm
Doktorandin

Timo Risch
Doktorand

Alexandra Amann
Technische Assistentin

Dr Carole Baumann
Technische Assistentin

Irene Kochems
Technische Assistentin

Karsten Mayr
Technischer Assistent

Kathrin Andres
Technische Assistentin

Mirjam Weil
Technische Assistentin

Nina Gampfer
Technische Assistentin

Sabine Backes
Technische Assistentin

Stefanie Neuber
Technische Assistentin

Verena Qallaku
Technische Assistentin

Viktoria George
Technische Assistentin

Besnik Qallaku
Laborhelfer
Forschungsprojekte
Isolierung neuer Myxobakterien für die Antibiotikaforschung
Metagenomische Analysen haben gezeigt, dass in der Natur eine Vielzahl an Myxobakterien existiert, die bislang noch nicht im Labor kultiviert werden konnten. Wir entwickeln Isolierungsstrategien um genau diese Stämme in Kultur zu bringen. Unsere Suche nach Myxobakterien umfasst die Analyse verschiedener Arten von Proben, Lebensräumen, und geographischen Lagen. Ein besonderer Fokus liegt auf bislang nur wenig erforschten Gebieten (z.B. Ozeane), sowie Habitaten mit einer hohen mikrobiellen Diversität und damit einhergehendem Konkurrenzdruck (z.B. Waldböden). Myxobakterien stellen eine vielversprechende Quelle für die Entdeckung bioaktiver Naturstoffe dar und unsere bisherigen Arbeiten haben gezeigt, dass die Art produzierten Naturstoffe sich stärker unterscheidet, je höher der phylogenetische Abstand zwischen zwei Stämmen ist (d.h. je weniger eng diese miteinander verwandt sind). Dementsprechend zielt unsere Isolierungsstrategie auf bislang noch nicht kultivierte Stämme ab, welche eine hohe Chance zur Entdeckung neuartiger Antiinfektiva bieten. Die Kultivierungsbedingungen werden im Labor gezielt auf die Bedingungen im Probenmaterial angepasst um das Bakterienwachstum und die Sekundärstoffproduktion zu unterstützen. Sobald ein Stamm erfolgreich isoliert wurde, folgt eine umfangreiche physiologische Charakterisierung, eine Optimierung der Wachstumsbedingungen, sowie Fermentationen um die Stämme auf bioaktive Substanzen hin untersuchen zu können. Weiterhin wird das Genom interessanter Stämme sequenziert um aufzuklären welche Gene für die Produktion eines bestimmten Stoffes verantwortlich sind, sowie weitere, bislang unbekannte Biosynthese-Gencluster zu identifizieren. In den letzten zwei Jahren haben wir zusätzlich damit begonnen, Myxobakterien aus Bodenproben von unterschiedlichen Teilen des Saarlandes zu isolieren. Interessierte Bürgerinnen und Bürger können das HIPS bei der Probennahme im Rahmen der Mitmach-Kampagne „Sample‘ das Saarland“ unterstützen.https://www.hips.saarland/sample/
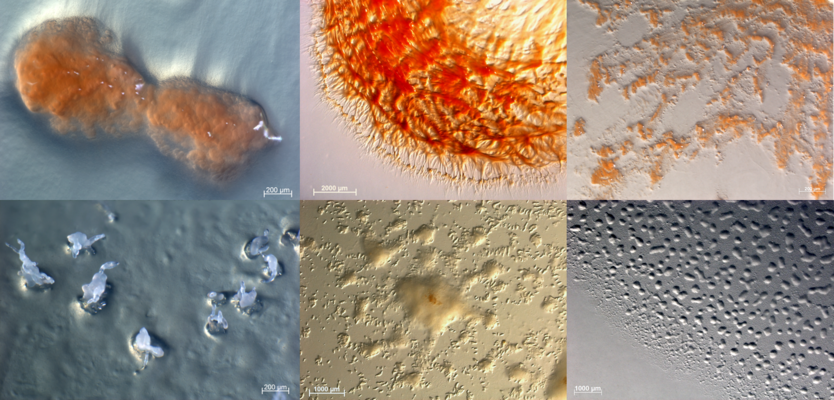
Neuartige Myxobakterien aus saarländischen Bodenproben
Cystobactamide als Breitbandantibiotika
Die Cystobactamide sind ein Beispiel für einen neuartigen, strukturell ungewöhnlichen Naturstoff aus Myxobakterien. Diese Gruppe nicht-ribosomal hergestellter Peptide setzt sich strukturell aus para-Aminobenzoesäure-Einheiten zusammen, welche zu einer Kette verknüpft und zusätzlich/anschließend modifiziert werden. Eine besondere Eigenschaft der Cystobactamide ist, dass sie sowohl auf grampositive als auch auf gramnegative Bakterien stark wachstumshemmend wirken. Vor allem gramnegative Erreger sind aufgrund ihrer doppelten Zellmembran üblicherweise besonders schwierig zu bekämpfen. Weiterhin zeigen die Cystobactamide keine Kreuzresistenzen mit gängigen Antibiotika wie den Fluorchinolonen. Die Kombination dieser Eigenschaften macht diese Substanzklasse zu potentiellen Kandidaten für den Einsatz als Breitbandantibiotika, weshalb sie in Kooperation mit dem Deutschen Zentrum für Infektionsforschung (DZIF) und seit Anfang 2019 auch mit der Firma Evotec weiterentwickelt werden. Das zelluläre Zielmolekül (Target) der Cystobactamide konnte mittels Analyse von Genomdaten resistenter Keime und nachfolgenden biochemischen Analysen aufgedeckt werden. Es handelt sich hierbei um die DNA-Gyrase, ein Enzym welches für die Überspiralisierung von DNA essentiell ist. Eine aus den Cystobactamiden abgeleitete Leitstruktur befindet sich mittlerweile in der präklinischen Entwicklungsphase und zeigte in Tiermodellen für bakterielle Infektionen sehr gute Aktivitäten. Damit ist der erste Schritt in der Entwicklung einer möglichen neuartigen Antibiotikaklasse getan.
Publikationen
2024
Isolation of Fidaxomicin and Shunt Metabolites from Actinoplanes deccanensis
Jung E, Hunter M, Dorst A, Major A, Teofilovic T, Müller R, Gademann K (2024)
HCADOI: 10.1002/hlca.202400013
The Peptide Antibiotic Corramycin Adopts a β-Hairpin-like Structure and Is Inactivated by the Kinase ComG
Adam S, Fries F, Tesmar A, Rasheed S, Deckarm S, Sousa C, Reberšek R, Risch T, Mancini S, Herrmann J, …, Kalinina O, Müller R (2024)
J. Am. Chem. Soc.DOI: 10.1021/jacs.3c13208
Nitroxoline resistance is associated with significant fitness loss and diminishes in vivo virulence of Escherichia coli
Deschner F, Risch T, Baier C, Schlüter D, Herrmann J, Müller R (2024)
Microbiology Spectrum 12 (1)DOI: 10.1128/spectrum.03079-23
New myxobacteria of the Myxococcaceae clade produce angiolams with antiparasitic activities
Walesch S, Garcia R, Mahmoud A, Panter F, Bollenbach S, Mäser P, Kaiser M, Krug D, Müller R (2024)
Microbiology SpectrumDOI: 10.1128/spectrum.03689-23
Mining the microbiota for antibiotics
Beemelmanns C, Keller A, Müller R (2024)
Nat. Microbiol. 9 (1): 13-14DOI: 10.1038/s41564-023-01568-8
Elucidation of unusual biosynthesis and DnaN-targeting mode of action of potent anti-tuberculosis antibiotics Mycoplanecins
Fu C, Liu Y, Walt C, Rasheed S, Bader C, Lukat P, Neuber M, Haeckl F, Blankenfeldt W, Kalinina O, Müller R (2024)
Nat Commun 15 (1)DOI: 10.1038/s41467-024-44953-5
2023
Inhibitors of the Elastase LasB for the Treatment of Pseudomonas aeruginosa Lung Infections
Konstantinovic J, Kany A, Alhayek A, Abdelsamie A, Sikandar A, Voos K, Yao Y, Andreas A, Shafiei R, Loretz B, …, Haupenthal J, Hirsch A (2023)
ACS Cent. Sci. 9 (12): 2205-2215DOI: 10.1021/acscentsci.3c01102
Molecular replacement for small-molecule crystal structure determination from X-ray and electron diffraction data with reduced resolution
Gorelik T, Lukat P, Kleeberg C, Blankenfeldt W, Mueller R (2023)
Acta crystallographica. Section A, Foundations and advances 79 (Pt 6): 504-514DOI: 10.1107/S2053273323008458
Darobactine: eine neue Antibiotikaklasse in Entwicklung
Seyfert C, Porten C, Müller R (2023)
Biospektrum 29 (5): 539-541DOI: 10.1007/s12268-023-1988-6
Tartrolon sensing and detoxification by the Listeria monocytogenes timABR resistance operon
Engelgeh T, Herrmann J, Jansen R, Müller R, Halbedel S (2023)
BookDOI: 10.1101/2023.07.27.550809
New Genetically Engineered Derivatives of Antibacterial Darobac-tins Underpin their Potential for Antibiotic Development
Seyfert C, Müller A, Walsh D, Birkelbach J, Kany A, Porten C, Yuan B, Krug D, Herrmann J, Marlovits T, Hirsch A, Müller R (2023)
BookDOI: 10.26434/chemrxiv-2023-24tmj
Impact of Drug Administration Routes on the In Vivo Efficacy of the Natural Product Sorangicin a Using a Staphylococcus aureus Infection Model in Zebrafish Embryos
Fries F, Kany A, Rasheed S, Hirsch A, Müller R, Herrmann J (2023)
International journal of molecular sciences 24 (16)DOI: 10.3390/ijms241612791
Optimization of Mass Spectrometry Imaging for Drug Metabolism and Distribution Studies in the Zebrafish Larvae Model: A Case Study with the Opioid Antagonist Naloxone
Park Y, Meyer M, Müller R, Herrmann J (2023)
International journal of molecular sciences 24 (12)DOI: 10.3390/ijms241210076
Revision of the Absolute Configurations of Chelocardin and Amidochelocardin
Sikandar A, Popoff A, Jumde R, Mándi A, Kaur A, Elgaher W, Rosenberger L, Hüttel S, Jansen R, Hunter M, …, Kurtán T, Müller R (2023)
Angewandte Chemie (International ed. in English)DOI: 10.1002/anie.202306437
Quantification of Plasmodium falciparum HRP-2 as an alternative method to 3Hhypoxanthine incorporation to measure the parasite reduction ratio in vitro
Carvalho L, Niepoth E, Mavraj-Husejni A, Kreidenweiss A, Herrmann J, Müller R, Knaab T, Burckhardt B, Kurz T, Held J (2023)
Int. J. Antimicrob. Agents 62 (3)DOI: 10.1016/j.ijantimicag.2023.106894
Schatztruhe Menschliches Mikrobiom: Wie Naturstoffe Uns Beeinflus...: Ingenta Connect
Schumm C, Donate P, Hegemann J, Keller A, Beemelmanns C, Müller R (2023)
Pharmakon 11 (4): 290-297DOI: 10.1691/pn.20230032
The Disorazole Z Family of Highly Potent Anticancer Natural Products from Sorangium cellulosum: Structure, Bioactivity, Biosynthesis, and Heterologous Expression
Gao Y, Birkelbach J, Fu C, Herrmann J, Irschik H, Morgenstern B, Hirschfelder K, Li R, Zhang Y, Jansen R, Müller R (2023)
Microbiology SpectrumDOI: 10.1128/spectrum.00730-23
Unusual peptide-binding proteins guide pyrroloindoline alkaloid formation in crocagin biosynthesis
Adam S, Zheng D, Klein A, Volz C, Mullen W, Shirran S, Smith B, Kalinina O, Müller R, Koehnke J (2023)
Nature chemistry 15 (4): 560-568DOI: 10.1038/s41557-023-01153-w
Insights into the biosynthesis of icumazole, unveiling a distinctive family of crotonyl-CoA carboxylase/reductase
Xie F, Kiefer A, Hirsch A, Kalinina O, Fu C, Müller R (2023)
Cell Reports Physical Science 4 (5)DOI: 10.1016/j.xcrp.2023.101394
Genome-Guided Discovery of the Myxobacterial Thiolactone-Containing Sorangibactins
Gao Y, Walt C, Bader C, Müller R (2023)
ACS chemical biology 18 (4): 924-932DOI: 10.1021/acschembio.3c00063
Evaluation of extraction methods for untargeted metabolomic studies for future applications in zebrafish larvae infection models
Schippers P, Rasheed S, Park Y, Risch T, Wagmann L, Hemmer S, Manier S, Müller R, Herrmann J, Meyer M (2023)
Sci. Rep. 13 (1)DOI: 10.1038/s41598-023-34593-y
Structure Elucidation and Biosynthesis of Nannosterols A and B, Myxobacterial Sterols from Nannocystis sp. MNa10993
Akone S, Hug J, Kaur A, Garcia R, Müller R (2023)
Journal of natural products 86 (4): 915-923DOI: 10.1021/acs.jnatprod.2c01143
The natural product chlorotonil A preserves colonization resistance and prevents relapsing Clostridioides difficile infection
Bublitz A, Brauer M, Wagner S, Hofer W, Müsken M, Deschner F, Lesker T, Neumann-Schaal M, Paul L, Nübel U, …, Fuchs T, Strowig T (2023)
Cell Host MicrobeDOI: 10.1016/j.chom.2023.04.003
The Myxobacterial Antibiotic Myxovalargin: Biosynthesis, Structural Revision, Total Synthesis, and Molecular Characterization of Ribosomal Inhibition
Koller T, Scheid U, Kösel T, Herrmann J, Krug D, Boshoff H, Beckert B, Evans J, Schlemmer J, Sloan B, …, Wilson D, Müller R (2023)
J. Am. Chem. Soc. 145 (2): 851-863DOI: 10.1021/jacs.2c08816
Fighting antibiotic resistance-strategies and (pre)clinical developments to find new antibacterials
Walesch S, Birkelbach J, Jézéquel G, Haeckl F, Hegemann J, Hesterkamp T, Hirsch A, Hammann P, Müller R (2023)
EMBO reports 24 (1)DOI: 10.15252/embr.202256033
2022
Total In Vitro Biosynthesis of the Thioamitide Thioholgamide and Investigation of the Pathway
Sikandar A, Lopatniuk M, Luzhetskyy A, Müller R, Koehnke J (2022)
J. Am. Chem. Soc. 144 (11): 5136-5144DOI: 10.1021/jacs.2c00402
Crystal structure of natural product argyrin-D determined by 3D electron diffraction
Gorelik T, Tehrani K, Gruene T, Monecke T, Niessing D, Kaiser U, Blankenfeldt W, Müller R (2022)
CrystEngComm 24 (33): 5885-5889DOI: 10.1039/D2CE00707J
Persicamidines-Unprecedented Sesquarterpenoids with Potent Antiviral Bioactivity against Coronaviruses
Keller L, Oueis E, Kaur A, Safaei N, Kirsch S, Gunesch A, Haid S, Rand U, Cicin-Šain L, Fu C, …, Pietschmann T, Müller R (2022)
Angewandte Chemie (International ed. in English)DOI: 10.1002/anie.202214595
Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering
Seyfert C, Porten C, Yuan B, Deckarm S, Panter F, Bader C, Coetzee J, Deschner F, Tehrani K, Higgins P, …, Herrmann J, Müller R (2022)
Angewandte Chemie (International ed. in English)DOI: 10.1002/anie.202214094
Current Developments in Antibiotic Discovery – Global Microbial Diversity as Source for Evolutionary Optimized Anti-infectives
Hegemann J, Birkelbach J, Walesch S, Müller R (2022)
EMBO Rep. published concurrently
Large-Scale Interlaboratory DI-FT-ICR MS Comparability Study Employing Various Systems
Forcisi S, Moritz F, Thompson C, Kanawati B, Uhl J, Afonso C, Bader C, Barsch A, Boughton B, Chu R, …, Laukien F, Schmitt-Kopplin P (2022)
Journal of the American Society for Mass SpectrometryDOI: 10.1021/jasms.2c00082
Total synthesis of Myxoprincomide, a secondary metabolite from Myxococcus xanthus
Kohr M, Walt C, Dastbaz J, Müller R, Kazmaier U (2022)
Org. Biomol. Chem.DOI: 10.1039/d2ob02021a
Pineapple Lectin AcmJRL binds SARS-CoV-2 Spike Protein in a carbohydrate-dependent fashion
Meiers J, Dastbaz J, Adam S, Rasheed S, Kirsch S, Meiser P, Gross P, Müller R, Titz A (2022)
Chembiochem : a European journal of chemical biologyDOI: 10.1002/cbic.202200463
Expanding the Ajudazol Cytotoxin Scaffold: Insights from Genome Mining, Biosynthetic Investigations, and Novel Derivatives
Zeng H, Birkelbach J, Hoffmann J, Popoff A, Volz C, Müller R (2022)
Journal of natural productsDOI: 10.1021/acs.jnatprod.2c00637
Discovery, Biosynthesis and Biological Activity of a Succinylated Myxochelin from the Myxobacterial strain MSr12020
Okoth D, Hug J, Garcia R, Müller R (2022)
BookDOI: 10.20944/preprints202209.0147.v1
Thiamyxins: Structure and Biosynthesis of Myxobacterial RNA-Virus Inhibitors
Haack P, Harmrolfs K, Bader C, Garcia R, Gunesch A, Haid S, Popoff A, Voltz A, Kim H, Bartenschlager R, Pietschmann T, Müller R (2022)
Angewandte Chemie (International ed. in English)DOI: 10.1002/anie.202212946
Structure Elucidation, Total Synthesis, Antibacterial In Vivo Efficacy and Biosynthesis Proposal of Myxobacterial Corramycin
Couturier C, Groß S, Tesmar A, Hoffmann J, Deckarm S, Fievet A, Dubarry N, Taillier T, Pöverlein C, Stump H, …, Müller R, Renard S (2022)
Angewandte Chemie (International ed. in English)DOI: 10.1002/anie.202210747
Systematic cross-biospecimen evaluation of DNA extraction kits for long- and short-read multi-metagenomic sequencing studies
Rehner J, Schmartz G, Groeger L, Dastbaz J, Ludwig N, Hannig M, Rupf S, Seitz B, Flockerzi E, Berger T, …, Keller A, Müller R (2022)
Genomics Proteomics BioinformaticsDOI: 10.1016/j.gpb.2022.05.006
Three New Stigmatellin Derivatives Reveal Biosynthetic Insights of Its Side Chain Decoration
Okoth D, Hug J, Garcia R, Müller R (2022)
Molecules (Basel, Switzerland) 27 (14)DOI: 10.3390/molecules27144656
Beyond the approved: target sites and inhibitors of bacterial RNA polymerase from bacteria and fungi
Kirsch S, Haeckl F, Müller R (2022)
Nat. Prod. Rep.DOI: 10.1039/d1np00067e
Regio- and Stereoselective Epoxidation and Acidic Epoxide Opening of Antibacterial and Antiplasmodial Chlorotonils Yield Highly Potent Derivatives
Müller R, Hofer W, Oueis E, Abou Fayad A, Deschner F, Andreas A, Carvalho L, Hüttel S, Bernecker S, Pätzold L, …, Held J, Herrmann J (2022)
Angewandte Chemie (International ed. in English)DOI: 10.1002/anie.202202816
Induction of Liver Size Reduction in Zebrafish Larvae by the Emerging Synthetic Cannabinoid 4F-MDMB-BINACA and Its Impact on Drug Metabolism
Park Y, Dahlem C, Meyer M, Kiemer A, Müller R, Herrmann J (2022)
Molecules (Basel, Switzerland) 27 (4)DOI: 10.3390/molecules27041290
Myxobacteria of the Cystobacterineae Suborder Are Producers of New Vitamin K2 Derived Myxoquinones
Panter F, Popoff A, Garcia R, Krug D, Müller R (2022)
Microorganisms 10 (3)DOI: 10.3390/microorganisms10030534
Novel 2,4-disubstituted quinazoline analogs as antibacterial agents with improved cytotoxicity profile: Modification of the benzenoid part
Megahed S, Rasheed S, Herrmann J, El-Hossary E, El-Shabrawy Y, Abadi A, Engel M, Müller R, Abdel-Halim M, Hamed M (2022)
Bioorg Med Chem Lett 59DOI: 10.1016/j.bmcl.2022.128531
New Deoxyenhygrolides from Plesiocystis pacifica Provide Insights into Butenolide Core Biosynthesis
Hug J, Kjaerulff L, Garcia R, Müller R (2022)
Marine drugs 20 (1)DOI: 10.3390/md20010072
Sandacrabins - Structurally Unique Antiviral RNA Polymerase Inhibitors from a Rare Myxobacterium
Bader C, Panter F, Garcia R, Tchesnokov E, Haid S, Walt C, Spröer C, Kiefer A, Götte M, Overmann J, Pietschmann T, Müller R (2022)
Chem. Eur. J. 28 (10)DOI: 10.1002/chem.202104484
Biotechnological production optimization of argyrins - a potent immunomodulatory natural product class
Pogorevc D, Müller R (2022)
Microbial Biotechnology 15 (1): 353-369DOI: 10.1111/1751-7915.13959
2021
Towards the sustainable discovery and development of new antibiotics
Miethke M, Pieroni M, Weber T, Brönstrup M, Hammann P, Halby L, Arimondo P, Glaser P, Aigle B, Bode H, …, Moser H, Müller R (2021)
Nature reviews. Chemistry 5 (10): 726-749DOI: 10.1038/s41570-021-00313-1
Genome-Guided Discovery of the First Myxobacterial Biarylitide Myxarylin Reveals Distinct C-N Biaryl Crosslinking in RiPP Biosynthesis
Hug J, Frank N, Walt C, Šenica P, Panter F, Müller R (2021)
Molecules 26 (24)DOI: 10.3390/molecules26247483
Structure and Biosynthesis of Myxofacyclines: Unique Myxobacterial Polyketides Featuring Varing and Rare Heterocycles *
Popoff A, Hug J, Walesch S, Garcia R, Keller L, Müller R (2021)
Chem. Eur. J. 27 (67): 16654-16661DOI: 10.1002/chem.202103095
Expanding the Myxochelin Natural Product Family by Nicotinic Acid Containing Congeners
Frank N, Széles M, Akone S, Rasheed S, Hüttel S, Frewert S, Hamed M, Herrmann J, Schuler S, Hirsch A, Müller R (2021)
Molecules 26 (16)DOI: 10.3390/molecules26164929
Improved broad-spectrum antibiotics against Gram-negative pathogens via darobactin biosynthetic pathway engineering
Groß S, Panter F, Pogorevc D, Seyfert C, Deckarm S, Bader C, Herrmann J, Müller R (2021)
Chem. Sci. 12 (35): 11882-11893DOI: 10.1039/d1sc02725e
Sesbanimide R, a Novel Cytotoxic Polyketide Produced by Magnetotactic Bacteria
Awal R, Haack P, Bader C, Riese C, Schüler D, Müller R (2021)
mBio 12 (3)DOI: 10.1128/mBio.00591-21
Zebrafish: An Attractive Model to Study Staphylococcus aureus Infection and Its Use as a Drug Discovery Tool
Rasheed S, Fries F, Müller R, Herrmann J (2021)
Pharmaceuticals 14 (6)DOI: 10.3390/ph14060594
Structure and biosynthesis of sorangipyranone - a new γ-dihydropyrone from the myxobacterial strain MSr12020
Okoth D, Hug J, Mándi A, Kurtán T, Garcia R, Müller R (2021)
Journal of Industrial Microbiology & Biotechnology 48 (3-4)DOI: 10.1093/jimb/kuab029
Search for the Active Ingredients from a 2-Aminothiazole DMSO Stock Solution with Antimalarial Activity
Ropponen H, Bader C, Diamanti E, Illarionov B, Rottmann M, Fischer M, Witschel M, Müller R, Hirsch A (2021)
ChemMedChem 16 (13): 2089-2093DOI: 10.1002/cmdc.202100067
Synergizing the potential of bacterial genomics and metabolomics to find novel antibiotics
Panter F, Bader C, Müller R (2021)
Chem. Sci. 12 (17): 5994-6010DOI: 10.1039/d0sc06919a
In vivo and in vitro reconstitution of unique key steps in cystobactamid antibiotic biosynthesis
Groß S, Schnell B, Haack P, Auerbach D, Müller R (2021)
Nat Commun 12 (1)DOI: 10.1038/s41467-021-21848-3
The Sandarazols are Cryptic and Structurally Unique Plasmid-Encoded Toxins from a Rare Myxobacterium*
Panter F, Bader C, Müller R (2021)
Angewandte Chemie (International ed. in English) 60 (15): 8081-8088DOI: 10.1002/anie.202014671
Natural products in drug discovery: advances and opportunities
Atanasov A, Zotchev S, Dirsch V, Supuran C (2021)
Nature reviews. Drug discovery 20 (3): 200-216DOI: 10.1038/s41573-020-00114-z
Expanding the Scope of Detectable Microbial Natural Products by Complementary Analytical Methods and Cultivation Systems
Bader C, Haack P, Panter F, Krug D, Müller R (2021)
Journal of natural products 84 (2): 268-277DOI: 10.1021/acs.jnatprod.0c00942
Kibdelosporangium persicum sp. nov., a new member of the Actinomycetes from a hot desert in Iran
Safaei N, Nouioui I, Mast Y, Zaburannyi N, Rohde M, Schumann P, Müller R, Wink J (2021)
Int. J. Syst. Evol. Microbiol. 71 (2)DOI: 10.1099/ijsem.0.004625
2020
Characterization of bromelain indicates a molar excess of inhibitor vs. enzyme molecules, a Jacalin-like lectin and Maillard reaction products
Gross P, Seelert H, Meiser P, Müller R (2020)
Journal of pharmaceutical and biomedical analysis 181DOI: 10.1016/j.jpba.2019.113075
The ROK like protein of Myxococcus xanthus DK1622 acts as a pleiotropic transcriptional regulator for secondary metabolism
Izzat S, Rachid S, Ajdidi A, El-Nakady Y, Liu X, Ye B, Müller R (2020)
J. Biotechnol. 311: 25-34DOI: 10.1016/j.jbiotec.2020.02.005
Human microbial metabolite mimicry as a strategy to expand the chemical space of potential drugs
Li H, Ranhotra H, Mani S, Dvorák Z, Sokol H, Müller R (2020)
Drug Discov. Today 25 (9): 1575-1579DOI: 10.1016/j.drudis.2020.06.007
Amidochelocardin Overcomes Resistance Mechanisms Exerted on Tetracyclines and Natural Chelocardin
Hennessen F, Miethke M, Zaburannyi N, Loose M, Lukežic T, Bernecker S, Hüttel S, Jansen R, Schmiedel J, Fritzenwanker M, …, Herrmann J, Müller R (2020)
Antibiotics 9 (9)DOI: 10.3390/antibiotics9090619
How to Study the Metabolism of New Psychoactive Substances for the Purpose of Toxicological Screenings-A Follow-Up Study Comparing Pooled Human Liver S9, HepaRG Cells, and Zebrafish Larvae
Wagmann L, Frankenfeld F, Park Y, Herrmann J, Fischmann S, Westphal F, Müller R, Flockerzi V, Meyer M (2020)
Frontiers in chemistry 8DOI: 10.3389/fchem.2020.00539
Natural Products Impacting DNA Methyltransferases and Histone Deacetylases
Akone S, Ntie-Kang F, Stuhldreier F, Ewonkem M, Noah A, Mouelle S, Müller R (2020)
Frontiers in pharmacology 11DOI: 10.3389/fphar.2020.00992
Heterologous expression of the atypical tetracycline chelocardin reveals the full set of genes required for its biosynthesis
Lukežic T, Pikl Š, Zaburannyi N, Remškar M, Petkovic H, Müller R (2020)
Microb. Cell Fact. 19 (1)DOI: 10.1186/s12934-020-01495-x
Corallopyronin A for short-course anti-wolbachial, macrofilaricidal treatment of filarial infections
Schiefer A, Hübner M, Krome A, Lämmer C, Ehrens A, Aden T, Koschel M, Neufeld H, Chaverra-Muñoz L, Jansen R, …, Pfarr K, Hoerauf A (2020)
PLoS Negl. Trop. Dis. 14 (12)DOI: 10.1371/journal.pntd.0008930
Leben und Überleben im Boden: Myxococcus xanthus ist Mikrobe des Jahres 2020
Volz C, Krug D, Müller R (2020)
Biol. Unserer Zeit 50 (6): 424-432DOI: 10.1002/biuz.202010721
Supercritical Fluid Extraction Enhances Discovery of Secondary Metabolites from Myxobacteria
Bader C, Neuber M, Panter F, Krug D, Müller R (2020)
Analytical chemistry 92 (23): 15403-15411DOI: 10.1021/acs.analchem.0c02995
An ambruticin-sensing complex modulates Myxococcus xanthus development and mediates myxobacterial interspecies communication
Marcos-Torres F, Volz C, Müller R (2020)
Nat Commun 11 (1)DOI: 10.1038/s41467-020-19384-7
Metabolic Profiling to Determine Bactericidal or Bacteriostatic Effects of New Natural Products using Isothermal Microcalorimetry
Cirnski K, Coetzee J, Herrmann J, Müller R (2020)
Journal of visualized experiments : JoVE (164)DOI: 10.3791/61703
Drug Administration Routes Impact the Metabolism of a Synthetic Cannabinoid in the Zebrafish Larvae Model
Park Y, Meyer M, Müller R, Herrmann J (2020)
Molecules 25 (19)DOI: 10.3390/molecules25194474
2-Hydroxysorangiadenosine: Structure and Biosynthesis of a Myxobacterial Sesquiterpene-Nucleoside
Okoth D, Hug J, Garcia R, Spröer C, Overmann J, Müller R (2020)
Molecules (Basel, Switzerland) 25 (11)DOI: 10.3390/molecules25112676
Biosynthesis of Cittilins, Unusual Ribosomally Synthesized and Post-translationally Modified Peptides from Myxococcus xanthus
Hug J, Dastbaz J, Adam S, Revermann O, Koehnke J, Krug D, Müller R (2020)
ACS chemical biology 15 (8): 2221-2231DOI: 10.1021/acschembio.0c00430
Bacteria as genetically programmable producers of bioactive natural products
Hug J, Krug D, Müller R (2020)
Nat. rev. chem. 4 (4): 172-193DOI: 10.1038/s41570-020-0176-1
Host Development for Heterologous Expression and Biosynthetic Studies of Myxobacterial Natural Products
Hug J, Müller R (2020)
In press: 149-216DOI: 10.1016/B978-0-12-409547-2.14818-8
Myxobakterielle Naturstofffabriken
Krug D, Garcia R, Müller R (2020)
Biospektrum 26 (1): 32-36DOI: 10.1007/s12268-020-1334-1
Dual-function chromogenic screening-based CRISPR/Cas9 genome editing system for actinomycetes
Wang Q, Xie F, Tong Y, Habisch R, Yang B, Zhang L, Müller R, Fu C (2020)
Applied Microbiology and Biotechnology 104 (1): 225-239DOI: 10.1007/s00253-019-10223-4
In depth natural product discovery - Myxobacterial strains that provided multiple secondary metabolites
Bader C, Panter F, Müller R (2020)
Biotechnol Adv 39DOI: 10.1016/j.biotechadv.2019.107480
2019
Ribosome-targeting antibiotics impair T cell effector function and ameliorate autoimmunity by blocking mitochondrial protein synthesis
Almeida L, Dhillon-LaBrooy A, Castro C, Ayele N, Bartel J, Carriche G, Guderian M, Lippens S, Dennerlein S, Hesse C, …, Moita L, Sparwasser T (2019)
Book 97DOI: 10.1101/832956
Clinical Resistome Screening of 1,110 Escherichia coli Isolates Efficiently Recovers Diagnostically Relevant Antibiotic Resistance Biomarkers and Potential Novel Resistance Mechanisms
Volz C, Ramoni J, Beisken S, Galata V, Keller A, Plum A, Posch A, Müller R (2019)
Front. Microbiol. 10DOI: 10.3389/fmicb.2019.01671
Polyunsaturated fatty acid production by Yarrowia lipolytica employing designed myxobacterial PUFA synthases
Gemperlein K, Dietrich D, Kohlstedt M, Zipf G, Bernauer H, Wittmann C, Wenzel S, Müller R (2019)
Nat. Commun. 10 (1)DOI: 10.1038/s41467-019-12025-8
Production of a Dibrominated Aromatic Secondary Metabolite by a Planctomycete Implies Complex Interaction with a Macroalgal Host
Panter F, Garcia R, Thewes A, Zaburannyi N, Bunk B, Overmann J, Gutierrez M, Krug D, Müller R (2019)
ACS Chem. Biol. 14 (accepted // 12): 2713-2719DOI: 10.1021/acschembio.9b00641
Production optimization and biosynthesis revision of corallopyronin A, a potent anti-filarial antibiotic
Pogorevc D, Panter F, Schillinger C, Jansen R, Wenzel S, Müller R (2019)
Metab. Eng. 55: 201-211DOI: 10.1016/j.ymben.2019.07.010
Two Biosynthetic Pathways in Jahnella thaxteri for Thaxteramides, Distinct Types of Lipopeptides
Oueis E, Klefisch T, Zaburannyi N, Garcia R, Plaza A, Müller R (2019)
Org. Lett. 21 (14): 5407-5412DOI: 10.1021/acs.orglett.9b01524
The role of protein-protein interactions in the biosynthesis of ribosomally synthesized and post-translationally modified peptides
Sikandar A, Koehnke J (2019)
Nat. Prod. Rep. 36 (11): 1576-1588DOI: 10.1039/C8NP00064F
Dedication: Heinz Floss and Christopher Walsh-pioneers in natural product chemical biology
Müller R, Wright G (2019)
Journal of Industrial Microbiology & Biotechnology 46 (3-4): 251-255DOI: 10.1007/s10295-019-02139-9
Engineering Atypical Tetracycline Formation in Amycolatopsis sulphurea for the Production of Modified Chelocardin Antibiotics
Lukežic T, Fayad A, Bader C, Harmrolfs K, Bartuli J, Groß S, Lešnik U, Hennessen F, Herrmann J, Pikl Š, Petkovic H, Müller R (2019)
ACS Chem. Biol. 14 (3): 468-477DOI: 10.1021/acschembio.8b01125
Armeniaspirol Antibiotic Biosynthesis: Chlorination and Oxidative Dechlorination Steps Affording Spiro4.4non-8-ene
Fu C, Xie F, Hoffmann J, Wang Q, Bauer A, Brönstrup M, Mahmud T, Müller R (2019)
ChemBioChem 20 (6): 764-769DOI: 10.1002/cbic.201800791
Heterologous expression of bacterial natural product biosynthetic pathways
Huo L, Hug J, Fu C, Bian X, Zhang Y, Müller R (2019)
Nat. Prod. Rep. 36: 1412-1436DOI: 10.1039/C8NP00091C
Genome mining reveals uncommon alkylpyrones as type III PKS products from myxobacteria
Hug J, Panter F, Krug D, Müller R (2019)
J. Ind. Microbiol. Biotechnol. 46 (3-4): 319-334DOI: 10.1007/s10295-018-2105-6
Class I Methyltransferase VioH Catalyzes Unusual S-Adenosyl-l-methionine Cyclization Leading to 4-Methylazetidinecarboxylic Acid Formation during Vioprolide Biosynthesis
Yan F, Müller R (2019)
ACS Chem. Biol. 14 (1): 99-105DOI: 10.1021/acschembio.8b00958
Novel Methoxymethacrylate Natural Products Uncovered by Statistics-Based Mining of the Myxococcus fulvus Secondary Metabolome
Panter F, Krug D, Müller R (2019)
ACS Chem. Biol. 14 (1): 88-98DOI: 10.1021/acschembio.8b00948
2018
Iterative Methylations Resulting in the Biosynthesis of the t-Butyl Group Catalyzed by a B12-Dependent Radical SAM Enzyme in Cystobactamid Biosynthesis
Wang Y, Schnell B, Müller R, Begley T (2018)
Methods in enzymology 606: 199-216DOI: 10.1016/bs.mie.2018.05.018
Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA
Sikandar A, Cirnski K, Testolin G, Volz C, Brönstrup M, Kalinina O, Müller R, Koehnke J (2018)
J. Am. Chem. Soc. 140 (48): 16641-16649DOI: 10.1021/jacs.8b08895
Characterization of an Unusual Glycerate Esterification Process in Vioprolide Biosynthesis
Auerbach D, Yan F, Zhang Y, Müller R (2018)
ACS Chem. Biol. 13 (11): 3123-3130DOI: 10.1021/acschembio.8b00826
Biocompatible bacteria-derived vesicles show inherent antimicrobial activity
Schulz E, Goes A, Garcia R, Panter F, Koch M, Müller R, Fuhrmann K, Fuhrmann G (2018)
Journal of Controlled Release 290: 46-55DOI: 10.1016/j.jconrel.2018.09.030
A polyphasic approach leads to seven new species of the cellulose-decomposing genus Sorangium, Sorangium ambruticinum sp. nov., Sorangium arenae sp. nov., Sorangium bulgaricum sp. nov., Sorangium dawidii sp. nov., Sorangium kenyense sp. nov., Sorangium orientale sp. nov. and Sorangium reichenbachii sp. nov
Mohr K, Wolf C, Nübel U, Szafranska A, Steglich M, Hennessen F, Gemperlein K, Kämpfer P, Martin K, Müller R, Wink J (2018)
Int. J. Syst. Evol. Microbiol. 68 (11)DOI: 10.1099/ijsem.0.003034
Activation of the NLRP3 inflammasome by hyaboron, a new asymmetric boron-containing macrodiolide from the Myxobacterium Hyalangium minutum
Surup F, Chauhan D, Niggemann J, Bartok E, Herrmann J, Koeck M, Zander W, Stadler M, Hornung V, Müller R (2018)
ACS Chem. Biol. 13 (10): 2981-2988DOI: 10.1021/acschembio.8b00659
Metabolic and Biosynthetic Diversity in Marine Myxobacteria
Gemperlein K, Zaburannyi N, Garcia R, La Clair J, Müller R (2018)
Mar. Drugs 16 (9)DOI: 10.3390/md16090314
Future Directions of Marine Myxobacterial Natural Product Discovery Inferred from Metagenomics
Garcia R, La Clair J, Müller R (2018)
Mar. Drugs 16 (9)DOI: 10.3390/md16090303
Simulacricoccus ruber gen. nov., sp. nov., a microaerotolerant, non-fruiting, myxospore-forming soil myxobacterium and emended description of the family Myxococcaceae
Garcia R, Müller R (2018)
Int. J. Syst. Evol. Microbiol. 68 (10): 3101-3110DOI: 10.1099/ijsem.0.002936
Structure, Total Synthesis, and Biosynthesis of Chloromyxamides: Myxobacterial Tetrapeptides Featuring an Uncommon 6-Chloromethyl-5-methoxypipecolic Acid Building Block
Gorges J, Panter F, Kjaerulff L, Hoffmann T, Kazmaier U, Müller R (2018)
Angew. Chem. Int. Ed. Engl. 57 (43): 14270-14275DOI: 10.1002/anie.201808028
Concepts and Methods to Access Novel Antibiotics from Actinomycetes
Hug J, Bader C, Remškar M, Cirnski K, Müller R (2018)
Antibiotics 7 (2)DOI: 10.3390/antibiotics7020044
Self-resistance guided genome mining uncovers new topoisomerase inhibitors from myxobacteria
Panter F, Krug D, Baumann S, Müller R (2018)
Chem. Sci. 9 (21): 4898-4908DOI: 10.1039/C8SC01325J
Biosynthesis and Heterologous Production of Vioprolides: Rational Biosynthetic Engineering and Unprecedented 4-Methylazetidinecarboxylic Acid Formation
Yan F, Auerbach D, Chai Y, Keller L, Tu Q, Hüttel S, Glemser A, Grab H, Bach T, Zhang Y, Müller R (2018)
Angew. Chem. Int. Ed. Engl. 57 (28): 8754-8759DOI: 10.1002/anie.201802479
Octapeptins: lipopeptide antibiotics against multidrug-resistant superbugs
Abou Fayad A, Herrmann J, Müller R (2018)
Cell Chem. Biol. 25 (4): 351-353DOI: 10.1016/j.chembiol.2018.04.003
Nannocystis konarekensis sp. nov., a novel myxobacterium from an Iranian desert
Mohr K, Moradi A, Glaeser S, Kämpfer P, Gemperlein K, Nübel U, Schumann P, Müller R, Wink J (2018)
Int. J. Syst. Evol. Microbiol. 68 (3): 721-729DOI: 10.1099/ijsem.0.002569
Biosynthesis of the Klebsiella oxytoca Pathogenicity Factor Tilivalline: Heterologous Expression, in Vitro Biosynthesis, and Inhibitor Development
Tesmar A, Hoffmann M, Abou Fayad A, Hüttel S, Schmitt V, Herrmann J, Müller R (2018)
ACS Chem. Biol. 13 (3): 812-819DOI: 10.1021/acschembio.7b00990
Crocadepsins-Depsipeptides from the Myxobacterium Chondromyces crocatus Found by a Genome Mining Approach
Surup F, Viehrig K, Rachid S, Plaza A, Maurer C, Hartmann R, Müller R (2018)
ACS Chem. Biol. 13 (1): 267-272DOI: 10.1021/acschembio.7b00900
Homospermidine Lipids: A compound class specifically formed during fruiting body formation of Myxococcus xanthus DK1622
Hoffmann M, Auerbach D, Panter F, Hoffmann T, Dorrestein P, Müller R (2018)
ACS Chem. Biol. 13 (1): 273-280DOI: 10.1021/acschembio.7b00816
Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria
Hoffmann T, Krug D, Bozkurt N, Duddela S, Jansen R, Garcia R, Gerth K, Steinmetz H, Müller R (2018)
Nat. Commun. 9 (1)DOI: 10.1038/s41467-018-03184-1
Production of myxopyronin and of its derivatives
Sucipto H, Sahner J, Wenzel S, Groh M, Hartmann R, Müller R (2018)
Patent (EP 2 994 535 B1)
2017
Novel and revisited approaches in antituberculosis drug discovery
Herrmann J, Rybniker J, Müller R (2017)
Current opinion in biotechnology 48: 94-101DOI: 10.1016/j.copbio.2017.03.023
Synthetic biology approaches to establish a heterologous production system for coronatines
Gemperlein K, Hoffmann M, Huo L, Pilak P, Petzke L, Müller R, Wenzel S (2017)
Metab. Eng. 44: 213-222DOI: 10.1016/j.ymben.2017.09.009
Heterologous production of myxobacterial α-pyrone antibiotics in Myxococcus xanthus
Sucipto H, Pogorevc D, Luxenburger E, Wenzel S, Müller R (2017)
Metab. Eng. 44: 160-170DOI: 10.1016/j.ymben.2017.10.004
Thioholgamides: Thioamide-Containing Cytotoxic RiPP Natural Products
Kjaerulff L, Sikandar A, Zaburannyi N, Adam S, Herrmann J, Koehnke J, Müller R (2017)
ACS Chem. Biol. 12 (11): 2837-2841DOI: 10.1021/acschembio.7b00676
Linoleic and palmitoleic acid block streptokinase-mediated plasminogen activation and reduce severity of invasive group A streptococcal infection
Rox K, Jansen R, Loof T, Gillen C, Bernecker S, Walker M, Chhatwal G, Müller R (2017)
Scientific Reports 7 (1)DOI: 10.1038/s41598-017-11276-z
Total Biosynthesis of the Pyrrolo4,2benzodiazepine Scaffold Tomaymycin on an In Vitro Reconstituted NRPS System
Tesmar A, Hoffmann M, Pippel J, Fayad A, Dausend-Werner S, Bauer A, Blankenfeldt W, Müller R (2017)
Cell Chem. Biol. 24 (10): 1216-1227DOI: 10.1016/j.chembiol.2017.08.001
Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity
Lukat P, Katsuyama Y, Wenzel S, Binz T, König C, Blankenfeldt W, Brönstrup M, Müller R (2017)
Chem. Sci. 8 (11)DOI: 10.1039/C7SC02622F
Total syntheses of cystobactamids and structural confirmation of cystobactamid 919-2
Trauner D, Cheng B, Müller R (2017)
Angew. Chem. Int. Ed. 56 (26): 7407-7410DOI: 10.1002/anie.201705387
Pyxipyrrolones: Structure elucidation and biosynthesis of cytotoxic myxobacterial metabolites
Kjaerulff L, Raju R, Panter F, Scheid U, Garcia R, Herrmann J, Müller R (2017)
Angew. Chem. Int. Ed. 56 (32): 9614-9618DOI: 10.1002/anie.201704790
The natural product carolacton inhibits folate-dependent C1 metabolism by targeting FolD/MTHFD
Fu C, Sikandar A, Donner J, Zaburannyi N, Herrmann J, Reck M, Wagner-Döbler I, Koehnke J, Müller R (2017)
Nat Commun 8 (1)DOI: 10.1038/s41467-017-01671-5
Vitiosangium cumulatum gen. nov., sp. nov. and Vitiosangium subalbum sp. nov., soil myxobacteria, and emended descriptions of the genera Archangium and Angiococcus, and of the family Cystobacteraceae
Awal R, Garcia R, Gemperlein K, Wink J, Kunwar B, Parajuli N, Müller R (2017)
Int. J. Syst. Evol. Microbiol. 67 (5): 1422-1430DOI: 10.1099/ijsem.0.001829
Disorazoles block group A streptococcal invasion into epithelial cells via interference with the host factor ezrin
Rox K, Rohde M, Chhatwal G, Müller R (2017)
Cell Chem. Biol. 24 (1): 159-170DOI: 10.1016/j.chembiol.2016.12.011
Structure and biosynthesis of crocagins: polycyclic postranslationally modified ribosomal peptides from Chondromyces crocatus
Viehrig K, Surup F, Volz C, Herrmann J, Abou Fayad A, Adam S, Kohnke J, Trauner D, Müller R (2017)
Angew. Chem. (56): 1-5DOI: 10.1002/anie.201612640
Solving the puzzle of the one-carbon loss in ripostatin biosynthesis
Fu C, Auerbach D, Li Y, Scheid U, Luxenburger E, Garcia R, Irschik H, Müller R (2017)
Angew. Chem. Int. Ed. Engl. 56 (8): 2192-2197DOI: 10.1002/anie.201609950
Natural products from myxobacteria: novel metabolites and bioactivities
Herrmann J, Fayad A, Müller R (2017)
Nat. Prod. Rep. 34 (2): 135-160DOI: 10.1039/C6NP00106H
2016
Racemicystis persica sp. nov., a novel myxobacterium from Iranian soil
Wink J, Moradi A, Ebrahimipour G, Mohr K, Kämpfer P, Glaeser S, Hennessen F, Gemperlein K, Awal R, Wolf C, Müller R (2016)
International Journal of Systematic and Evolutionary Microbiology epub ahead of printDOI: 10.1099/ijsem.0.001655
Heterologous expression of the plant cysteine protease bromelain and its inhibitor in Pichia pastoris
Luniak N, Meiser P, Burkart S, Müller R (2016)
Biotechnol Progress 33 (1): 54-65DOI: 10.1002/btpr.2405
Strategies for the discovery and development of new antibiotics from natural products: Three case studies
Herrmann J, Lukezic T, Kling A, Baumann S, Hüttel S, Petkovic H, Müller R (2016)
Contribution: 1-25DOI: 10.1007/82_2016_498
Structure and Substrate Recognition of the Bottromycin Maturation Enzyme BotP
Mann G, Huo L, Adam S, Nardone B, Vendome J, Westwood N, Muller R, Koehnke J (2016)
ChemBioChem 17 (23): 2286-2292DOI: 10.1002/cbic.201600406
Discovery of the first small-molecule CsrA-RNA interaction inhibitors using biophysical screening technologies
Maurer C, Fruth M, Empting M, Avrutina O, Hossmann J, Nadmid S, Gorges J, Herrmann J, Kazmaier U, Dersch P, Muller R, Hartmann R (2016)
Future medicinal chemistry 8 (9): 931-47DOI: 10.4155/fmc-2016-0033
Isolation, structure elucidation, and (bio)synthesis of Haprolid, a cell-type-specific myxobacterial cytotoxin
Steinmetz H, Li J, Fu C, Zaburannyi N, Kunze B, Harmrolfs K, Schmitt V, Herrmann J, Reichenbach H, Hofle G, Kalesse M, Müller R (2016)
Angew. Chem. Int. Ed. Engl. 55 (34): 10113-7DOI: 10.1002/anie.201603288
Predicting the presence of uncommon elements in unknown biomolecules from isotope patterns
Meusel M, Hufsky F, Panter F, Krug D, Müller R, Böcker S (2016)
Anal. Chem. 88: 7556-7566DOI: 10.1021/acs.analchem.6b01015
Room temperature electrocompetent bacterial cells improve DNA transformation and recombineering efficiency
Tu Q, Yin J, Fu J, Herrmann J, Li Y, Yin Y, Stewart A, Müller R, Zhang Y (2016)
Sci. Rep. 6DOI: 10.1038/srep24648
Racemicystis crocea gen. nov., sp. nov., a novel soil myxobacterium in Polyangiaceae
Awal R, Garcia R, Müller R (2016)
Int. J. Syst. Evol. Microbiol. (66)DOI: 10.1099/ijsem.0.001045
RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression
Wang H, Li Z, Jia R, Hou Y, Yin J, Bian X, Li A, Muller R, Stewart A, Fu J, Zhang Y (2016)
Nat. Protoc. 11 (7): 1175-1190DOI: 10.1038/nprot.2016.054
Genetic engineering and heterologous expression of the disorazol biosynthetic gene cluster via Red/ET recombineering
Tu Q, Herrmann J, Hu S, Raju R, Bian X, Zhang Y, Müller R (2016)
Sci. Rep. 6DOI: 10.1038/srep21066
Genome analysis of the fruiting body forming myxobacterium Chondromyces crocatus reveals high potential for natural product Biosynthesis
Zaburannyi N, Bunk B, Maier J, Overmann J, Müller R (2016)
Appl. Environ. Microbiol. 82 (6): 1945-57DOI: 10.1128/AEM.03011-15
Investigations to the antibacterial mechanism of action of kendomycin
Elnakady Y, Chatterjee I, Bischoff M, Rohde M, Josten M, Sahl H, Herrmann M, Müller R (2016)
PLoS ONE 11 (1): 1-18DOI: 10.1371/journal.pone.0146165
Aetherobacter fasciculatus gen. nov., sp. nov. and Aetherobacter rufus gen. nov., sp. nov., two novel myxobacteria with promising biotechnological applications
Garcia R, Stadler M, Gemperlein K, Müller R (2016)
Int. J. Syst. Evol. Microbiol. 66: 928-938DOI: 10.1099/ijsem.0.000813
Metabolic engineering of Pseudomonas putida for production of docosahexaenoic acid based on a myxobacterial PUFA synthase
Gemperlein K, Zipf G, Bernauer H, Müller R, Wenzel S (2016)
Metab. Eng. 33: 98-108DOI: 10.1016/j.ymben.2015.11.001
2015
Synthetic pathway enzymes for the production of Argyrins
Müller R, Wenzel S, Garcia R (2015)
Patent (US9217169 B2)
Heterocyclyl-Butanamide Derivatives
Buchstaller H, Dorsch D (2015)
Patent (WO/2015/169421)
Two of a kind-The biosynthetic pathways of chlorotonil and anthracimycin
Jungmann K, Jansen R, Gerth K, Huch V, Krug D, Fenical W, Müller R (2015)
ACS Chem. Biol. 10 (11): 2480-2490DOI: 10.1021/acschembio.5b00523
Direct cloning and heterologous expression of the salinomycin biosynthetic gene cluster from Streptomyces albus DSM41398 in Streptomyces coelicolor A3(2)
Yin J, Hoffmann M, Bian X, Tu Q, Yan F, Xia L, Ding X, Stewart A, Müller R, Fu J, Zhang Y (2015)
Sci. Rep. 5DOI: 10.1038/srep15081
Production of the bengamide class of marine natural products in myxobacteria: biosynthesis and structure-activity relationships
Wenzel S, Hoffmann H, Zhang J, Debussche L, Haag-Richter S, Kurz M, Nardi F, Lukat P, Kochems I, Tietgen H, …, Müller R, Brönstrup M (2015)
Angew. Chem. Int. Ed. Engl. 54 (51): 15560-4DOI: 10.1002/anie.201508277
Construction of a new class of tetracycline lead structures with potent antibacterial activity through biosynthetic engineering
Lešnik U, Lukežic T, Podgoršek A, Horvat J, Polak T, Šala M, Jenko B, Harmrolfs K, Ocampo-Sosa A, Martínez-Martínez L, …, Müller R, Petkovic H (2015)
Angew. Chem. Int. Ed. Engl. 54 (13): 3937-3940DOI: 10.1002/anie.201411028
Pinensins: the first antifungal lantibiotics
Mohr K, Volz C, Jansen R, Wray V, Hoffmann J, Bernecker S, Wink J, Gerth K, Stadler M, Müller R (2015)
Angew. Chem. Int. Ed. Engl. 127 (38): 11254-11258DOI: 10.1002/anie.201500927
Cystochromones, unusual chromone-containing polyketides from the myxobacterium Cystobacter sp. MCy9104
Nadmid S, Plaza A, Garcia R, Müller R (2015)
J. Nat. Prod. 78 (8): 2023-8DOI: 10.1021/acs.jnatprod.5b00343
Macyranones: Structure, Biosynthesis, and Binding Mode of an Unprecedented Epoxyketone that Targets the 20S Proteasome
Keller L, Plaza A, Dubiella C, Groll M, Kaiser M, Müller R (2015)
J. Am. Chem. Soc. 137 (25): 8121-30DOI: 10.1021/jacs.5b03833
Biosynthetic studies of telomycin reveal new lipopeptides with enhanced activity
Fu C, Keller L, Bauer A, Brönstrup M, Froidbise A, Hammann P, Herrmann J, Mondesert G, Kurz M, Schiell M, …, Wink J, Müller R (2015)
J. Am. Chem. Soc. 137 (24): 7692-7705DOI: 10.1021/jacs.5b01794
Targeting DnaN for tuberculosis therapy using novel griselimycins
Kling A, Lukat P, Almeida D, Bauer A, Fontaine E, Sordello S, Zaburannyi N, Herrmann J, Wenzel S, König C, …, Lagrange S, Müller R (2015)
Science 348 (6239): 1106-1112DOI: 10.1126/science.aaa4690
In Vitro reconstitution of α-Pyrone ring formation in myxopyronin biosynthesis
Sucipto H, Sahner J, Prusov E, Wenzel S, Hartmann R, Koehnke J, Müller R (2015)
Chem. Sci. 6: 5076-5085DOI: 10.1039/C5SC01013F
Two more pieces of the colibactin genotoxin puzzle from Escherichia coli show incorporation of an unusual 1-aminocyclopropanecarboxylic acid moiety
Bian X, Plaza A, Zhang Y, Müller R (2015)
Chem. Sci. 6 (5): 3154-3160DOI: 10.1039/c5sc00101c
Synthesis and biological evaluation of cystobactamid 507: a bacterial topoisomerase inhibitor from Cystobacter sp
Moreno M, Elgaher W, Herrmann J, Schläger N, Hamed M, Baumann S, Müller R, Hartmann R, Kirschning A (2015)
Synlett 26 (9): 1175-1178DOI: 10.1055/s-0034-1380509
Rational and efficient site-directed mutagenesis of adenylation domain alters relative yields of luminmide derivatives in vivo
Bian X, Plaza A, Yan F, Zhang Y, Müller R (2015)
Biotechnol. Bioeng. 112 (7): 1343-53DOI: 10.1002/bit.25560
Advanced mutasynthesis studies on the natural α-pyrone antibiotic myxopyronin from Myxococcus fulvus
Sahner J, Sucipto H, Wenzel S, Groh M, Hartmann R, Müller R (2015)
ChemBioChem 16: 946-953DOI: 10.1002/cbic.201402666
Cystodienoic acid: A new diterpene isolated from the myxobacterium Cystobacter sp.
Raju R, Mohr K, Bernecker S, Herrmann J, Müller R (2015)
J. Antibiot. 68: 473-475DOI: 10.1038/ja.2015.8
Heterologous expression of an orphan NRPS gene cluster from Paenibacillus larvae in Escherichia coli revealed production of sevadicin
Tang Y, Frewert S, Harmrolfs K, Herrmann J, Karmann L, Kazmaier U, Xia L, Zhang Y, Müller R (2015)
J. Biotechnol. 194: 112-114DOI: 10.1016/j.jbiotec.2014.12.008
Aggregicoccus edonensis gen. nov., sp. nov., an unusually aggregating myxobacterium isolated from a soil sample
Sood S, Awal R, Wink J, Mohr K, Rohde M, Stadler M, Kämpfer P, Glaeser S, Schumann P, Garcia R, Müller R (2015)
Int. J. Syst. Evol. Microbiol. 65 (Pt 3): 745-753DOI: 10.1099/ijs.0.061176-0
2014
Cystobactamids: myxobacterial topoisomerase inhibitors exhibiting potent antibacterial activity
Baumann S, Herrmann J, Raju R, Steinmetz H, Mohr K, Hüttel S, Harmrolfs K, Stadler M, Müller R (2014)
Angew. Chem. Int. Ed. 53 (52): 14605-14609DOI: 10.1002/anie.201409964
Nannozinones and Sorazinones, Unprecedented Pyrazinones from Myxobacteria
Jansen R, Sood S, Mohr K, Kunze B, Irschik H, Stadler M, Müller R (2014)
J. Nat. Prod. 77 (11): 2545-2552DOI: 10.1021/np500632c
Disciformycins A and B: 12-membered macrolide glycoside antibiotics from the myxobacterium Pyxidicoccus fallax active against multiresistant staphylococci
Surup F, Viehrig K, Mohr K, Herrmann J, Jansen R, Müller R (2014)
Angew. Chem. Int. Ed. Engl. 49 (53): 13588-13591DOI: 10.1002/anie.201406973
Albaflavenol B, a new sesquiterpene isolated from the terrestrial actinomycete, Streptomyces sp.
Raju R, Gromyko O, Fedorenko V, Luzhetskyy A, Müller R (2014)
J. Antibiot. 68 (6): 286-288DOI: 10.1038/ja.2014.138
Improving natural products identification through targeted LC-MS/MS in an untargeted secondary metabolomics workflow
Hoffmann T, Krug D, Hüttel S, Müller R (2014)
Anal. Chem. 86 (21): 10780-10788DOI: 10.1021/ac502805w
Minicystis rosea gen. nov., sp. nov., a polyunsaturated fatty acid-rich and steroid-producing soil myxobacterium
Garcia R, Gemperlein K, Müller R (2014)
Int. J. Syst. Evol. Microbiol. 64 (Pt 11): 3733-3742DOI: 10.1099/ijs.0.068270-0
Cystobactamid und Derivate als Antibiotika
Baumann S, Herrmann J, Mohr K, Steinmetz H, Gerth K, Raju R, Müller R (2014)
Patent (1451)
Antimalarial activity of the myxobacterial macrolide chlorotonil A
Held J, Gebru T, Kalesse M, Jansen R, Gerth K, Müller R, Mordmüller B (2014)
Antimicrob. Agents Chemother. 58 (11): 6378-6384DOI: 10.1128/AAC.03326-14
Paenilarvins: Iturin family lipopeptides from the honey bee pathogen Paenibacillus larvae
Sood S, Steinmetz H, Beims H, Mohr K, Stadler M, Djukic M, Ohe W, Steinert M, Daniel R, Müller R (2014)
ChemBioChem 15 (13): 1947-1955DOI: 10.1002/cbic.201402139
Heterologous production of glidobactins/luminmycins in Escherichia coli nissle containing the glidobactin biosynthetic gene cluster from Burkholderia DSM7029
Bian X, Huang F, Wang H, Klefisch T, Müller R, Zhang Y (2014)
ChemBioChem 15 (15): 2221-2224DOI: 10.1002/cbic.201402199
Targeting the actin cytoskeleton: selective anti-tumor action via trapping PKCå
Vollmar A, Förster F, Braig S, Moser C, Kubisch R, Busse J, Wagner E, Schmöckel E, Mayr D, Schmitt S, …, Zischka H, Müller R (2014)
Cell Death & Disease 5DOI: 10.1038/cddis.2014.363
A highly unusual polyketide synthase directs dawenol polyene biosynthesis in Stigmatella aurantiaca
Oßwald C, Zaburannyi N, Burgard C, Hoffmann T, Wenzel S, Müller R (2014)
J. Biotechnol. 191: 54-63DOI: 10.1016/j.jbiotec.2014.07.447
Hyalachelins A-C, unusual siderophores isolated from the terrestrial myxobacterium Hyalangium minutum
Nadmid S, Plaza A, Lauro G, Garcia R, Bifulco G, Müller R (2014)
Org. Lett. 16 (16): 4130-4133DOI: 10.1021/ol501826a
Hyafurones, hyapyrrolines, and hyapyrones: polyketides from Hyalangium minutum
Okanya P, Mohr K, Gerth K, Kessler W, Jansen R, Stadler M, Müller R (2014)
J. Nat. Prod. 77 (6): 1420-1429DOI: 10.1021/np500145f
Biosynthesis of crocacin involves an unusual hydrolytic release domain showing similarity to condensation domains
Müller S, Rachid S, Hoffmann T, Surup F, Volz C, Zaburannyi N, Müller R (2014)
Chem. Biol. 21 (7): 855-865DOI: 10.1016/j.chembiol.2014.05.012
Angiolactone, a new Butyrolactone isolated from the terrestrial myxobacterium, Angiococcus sp.
Raju R, Garcia R, Müller R (2014)
J. Antibiot. epub ahead of printDOI: 10.1038/ja.2014.55
Cystomanamides: structure and biosynthetic pathway of a family of glycosylated lipopeptides from myxobacteria
Etzbach L, Plaza A, Garcia R, Baumann S, Müller R (2014)
Org. Lett. 16 (9): 2414-2417DOI: 10.1021/ol500779s
Indothiazinone, an indolyl-thiazolyl-ketone from a novel myxobacterium belonging to the Sorangiineae
Jansen R, Mohr K, Bernecker S, Stadler M, Müller R (2014)
J. Nat. Prod. 77 (4): 1054-1060DOI: 10.1021/np500144t
Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products
Krug D, Müller R (2014)
Nat. Prod. Rep. 31 (6): 768-783DOI: 10.1039/c3np70127a
Pyrronazols, metabolites from the myxobacteria Nannocystis pusilla and N. exedens, are unusual chlorinated pyrone-oxazole-pyrroles
Jansen R, Sood S, Huch V, Kunze B, Stadler M, Müller R (2014)
J. Nat. Prod. 77 (2): 320-326DOI: 10.1021/np400877r
Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters
Kolinko I, Lohße A, Borg S, Raschdorf R, Jogler C, Tu Q, Pósfai M, Tompa E, Plitzko J, Brachmann A, …, Zhang Y, Schüler D (2014)
Nat. Nanotechnol. 9 (3): 193-197DOI: 10.1038/nnano.2014.13
Oleamycins A and B: new antibacterial cyclic hexadepsipeptides isolated from a terrestrial Streptomyces sp.
Raju R, Gromyko O, Andriy B, Fedorenko V, Luzhetskyy A, Müller R (2014)
Journal of Antibiotics (Tokyo) 67 (4): 339-343DOI: 10.1038/ja.2014.1
Polyunsaturated fatty acid biosynthesis in myxobacteria
Gemperlein K, Rachid S, Garcia R, Wenzel S, Müller R (2014)
Chem. Sci. 5 (5): 1733-1741DOI: 10.1039/C3SC53163E
Improved seamless mutagenesis by recombineering using ccdB for counterselection
Wang H, Bian X, Xia L, Ding X, Müller R, Zhang Y, Fu J, Stewart A (2014)
Nucleic Acids Res. 42 (5)DOI: 10.1093/nar/gkt1339
Modular Construction of a Functional Artificial Epothilone Polyketide Pathway
Oßwald C, Zipf G, Schmidt G, Maier J, Bernauer H, Müller R, Wenzel S (2014)
ACS Synth. Biol. 3 (10): 759-772DOI: 10.1021/sb300080t
Future potential for anti-infectives from bacteria - How to exploit biodiversity and genomic potential
Müller R, Wink J (2014)
Int. J. Med. Microbiol. 304 (1): 3-13DOI: 10.1016/j.ijmm.2013.09.004
2013
Novel macrolide antibiotics
Müller R, Nett M, Surup F, Steinmetz H, Mohr K, Viehrig K (2013)
Patent (EP14002319)
Jahnellamides, a-keto-ß-methionine-containing peptides from the terrestrial myxobacterium Jahnella sp.: structure and biosynthesis
Plaza A, Viehrig K, Garcia R, Müller R (2013)
Org. Lett. 15 (22): 5882-5885DOI: 10.1021/ol402967y
Microsclerodermins from terrestrial myxobacteria: An intriguing biosynthesis likely connected to a sponge symbiont
Hoffmann T, Müller S, Nadmid S, Garcia R, Müller R (2013)
J. Am. Chem. Soc. 45 (135): 16904-16911DOI: 10.1021/ja4054509
Analytics of the therapeutic peptide aviptadil by sheathless CE-MS and comparison with nanoRP-HPLC-MS
Groß P, Burkart S, Müller R (2013)
J. Pharm. Biomed. Anal. 88 (2014): 477-482DOI: 10.1016/j.jpba.2013.09.024
Concerted action of P450 plus helper protein to form the amino-hydroxy-piperidone moiety of the potent protease inhibitor crocapeptin
Viehrig K, Surup F, Harmrolfs K, Jansen R, Kunze B, Müller R (2013)
J. Am. Chem. Soc. 135 (45): 16885-16894DOI: 10.1021/ja4047153
Production of fatty acids by heterologous expression of gene clusters from myxobacteria
Wenzel S, Rachid S, Gemperlein K, Müller R (2013)
Patent AC12P764FI (WO 2011/151298 A1)
Discovery and biological activity of new chondramides from Chondromyces sp.
Herrmann J, Hüttel S, Müller R (2013)
ChemBioChem 14 (13): 1573-1580DOI: 10.1002/cbic.201300140
Synthesis and biological activities of the respiratory chain inhibitor aurachin D and new ring versus chain analogues
Li X, Herrmann J, Zang Y, Grellier P, Prado S, Müller R, Nay B (2013)
Beilstein J. Org. Chem. 9: 1551-1558DOI: 10.3762/bjoc.9.176
Oleaceran: A novel spiro[isobenzofuran-1,2'-naptho[1,8-bc]furan] isolated from a terrestrial Streptomyces sp
Raju R, Gromyko O, Fedorenko V, Luzhetskyy A, Müller R (2013)
Org. Lett. 15 (14): 3487-3489DOI: 10.1021/ol401490u
Recent advances in the heterologous expression of microbial natural product biosynthetic pathways
Ongley S, Bian X, Neilan B, Müller R (2013)
Nat. Prod. Rep. 30 (8): 1121-1138DOI: 10.1039/c3np70034h
In vivo evidence for a prodrug activation mechanism during colibactin maturation
Bian X, Fu J, Plaza A, Herrmann J, Pistorius D, Stewart A, Zhang Y, Müller R (2013)
ChemBioChem 14 (10): 1194-1197DOI: 10.1002/cbic.201300208
Lorneic acids C and D, new trialkyl-substituted aromatic acids isolated from a terrestrial Streptomyces sp.
Raju R, Gromyko O, Fedorenko V, Luzhetskyy A, Müller R (2013)
J. Antibiot. 66 (6): 347-349DOI: 10.1038/ja.2013.18
Isolation, structure elucidation, and biological activity of maltepolides: remarkable macrolides from myxobacteria
Irschik H, Washausen P, Sasse F, Fohrer J, Huch V, Müller R, Prusov E (2013)
Angew. Chem. Int. Ed. Engl. 52 (20): 5402-5405DOI: 10.1002/anie.201210113
Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature
Arnison P, Bibb M, Bierbaum G, Bowers A, Bugni T, Bulaj G, Camarero J, Campopiano D, Challis G, Clardy J, …, Willey J, van der Donk W (2013)
Nat. Prod. Rep. 30 (1): 108-160DOI: 10.1039/C2NP20085F
Activity-guided screening of bioactive natural compounds implementing a new glucocorticoid-receptor-translocation assay and detection of new anti-inflammatory steroids from bacteria
Kaufmann K, Simmons L, Herrmann J, Schwär G, Luniak N, Müller R (2013)
Biotechnol. Lett. 35 (1): 11-20DOI: 10.1007/s10529-012-1042-0
2012
Method for producing recombinant 11-de-O-methyltomaymycin
Lattemann C, Broenstrup M, Werner S, Müller R, Harmrolfs K (2012)
Patent 2012 (EP2849753)
Pellasoren: structure elucidation, biosynthesis, and total synthesis of a cytotoxic secondary metabolite from Sorangium cellulosum
Jahns C, Hoffmann T, Müller S, Gerth K, Washausen P, Höfle G, Reichenbach H, Kalesse M, Müller R (2012)
Angew. Chem. Int. Ed. Engl. 51 (21): 5239-5243DOI: 10.1002/anie.201200327
Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting
Fu J, Bian X, Hu S, Wang H, Huang F, Seibert P, Plaza A, Xia L, Müller R, Stewart A, Zhang Y (2012)
Nat. Biotechnol. 30 (5): 440-446DOI: 10.1038/nbt.2183
Rubimycinone A, a new anthraquinone from a terrestrial Streptomyces sp.
Raju R, Gromyko O, Fedorenko V, Herrmann J, Luzhetskyy A, Müller R (2012)
Tetrahedron Lett. 54 (8): 900-902DOI: 10.1016/j.tetlet.2012.11.130
Ein alternativer isovaleryl-CoA-biosyntheseweg: beteiligung einer bisher unbekannten 3-methylglutaconyl-CoA-decarboxylase
Li Y, Luxenburger E, Müller R (2012)
Angew. Chem.DOI: 10.1002/ange.201207984
Juniperolide A: a new polyketide isolated from a terrestrial actinomycete, Streptomyces sp.
Raju R, Gromyko O, Fedorenko V, Luzhetskyy A, Plaza A, Müller R (2012)
Org. Lett. 14 (23): 5860-5863DOI: 10.1021/ol302766z
An alternative isovaleryl CoA biosynthetic pathway involving a previously unknown 3-methylglutaconyl CoA decarboxylase
Li Y, Luxenburger E, Müller R (2012)
Angew. Chem. Int. Ed. Engl. 52 (4): 1304-1308DOI: 10.1002/anie.201207984
Enhancer binding proteins act as hetero-oligomers and link secondary metabolite production to myxococcal development, motility and predation
Volz C, Kegler C, Müller R (2012)
Chem. Biol. 19: 1447-1459DOI: 10.1016/j.chembiol.2012.09.010
Indiacens A and B: prenyl indoles from the myxobacterium Sandaracinus amylolyticus
Steinmetz H, Mohr K, Zander W, Jansen R, Gerth K, Müller R (2012)
J. Nat. Prod. 75 (10): 1803-1805DOI: 10.1021/np300288b
Synthetic biotechnology to study and engineer ribosomal bottromycin biosynthesis
Huo L, Rachid S, Stadler M, Wenzel S, Müller R (2012)
Chem. Biol. 19 (10): 1278-1287DOI: 10.1016/j.chembiol.2012.08.013
Leopolic acid A, isolated from a terrestrial actinomycete, Streptomyces sp.
Raju R, Gromyko O, Fedorenko V, Luzhetskyy A, Müller R (2012)
Tetrahedron Lett. 53 (46): 6300-6301DOI: 10.1016/j.tetlet.2012.09.046
Aetheramides A and B, potent HIV-inhibitory depsipeptides from a myxobacterium of the new genus 'Aetherobacter'
Plaza A, Garcia R, Bifulco G, Martinez J, Hüttel S, Sasse F, Meyerhans A, Stadler M, Müller R (2012)
Org. Lett. 14 (11): 2854-2857DOI: 10.1021/ol3011002
Methods to optimize myxobacterial fermentations using off-gas analysis
Hüttel S, Müller R (2012)
Microb. Cell Fact. 11 (59)DOI: 10.1186/1475-2859-11-59
Pretubulysin: From hypothetical biosynthetic intermediate to potential lead in tumor therapy
Herrmann J, Elnakady Y, Wiedmann R, Ullrich A, Rohde M, Kazmaier U, Vollmar A, Müller R (2012)
PLoS ONE 7 (5)DOI: 10.1371/journal.pone.0037416
Luminmycins A-C, cryptic natural products from Photorhabdus luminescens identified by heterologous expression in Escherichia coli
Bian X, Plaza A, Zhang Y, Müller R (2012)
J. Nat. Prod. 75 (9): 1652-1655DOI: 10.1021/np300444e
Precursor-directed syntheses and biological evaluation of new elansolid derivatives
Steinmetz H, Zander W, Shushni M, Jansen G, Dehn R, Dräger G, Kirschning A, Müller R (2012)
ChemBioChem 13 (12): 1813-1817DOI: 10.1002/cbic.201200228
Direct cloning, genetic engineering, and heterologous expression of the syringolin biosynthetic gene cluster in E. coli through Red/ET recombineering
Bian X, Huang F, Stewart F, Xia L, Zhang Y, Müller R (2012)
ChemBioChem 13 (13): 1946-1952DOI: 10.1002/cbic.201200310
Eine Semipinakol-Umlagerung - katalysiert durch ein Enzymsystem mit difunktioneller FAD-abhängiger Monooxygenase
Katsuyama Y, Harmrolfs K, Pistorius D, Li Y, Müller R (2012)
Angew. Chem. Int. Ed. Engl. 124 (37): 9573-9576DOI: 10.1002/ange.201204138
A semipinacol rearrangement directed by an enzymatic system featuring dual-function FAD-dependent monooxygenase
Katsuyama Y, Harmrolfs K, Pistorius D, Li Y, Müller R (2012)
Angew. Chem. Int. Ed. Engl. 51 (37): 9437-9440DOI: 10.1002/anie.201204138
Pimprinols A-C, from the terrestrial actinomycete, Streptomyces sp.
Raju R, Gromyko O, Fedorenko V, Luzhetskyy A, Müller R (2012)
Tetrahedron Lett. 53 (24): 3009-3011DOI: 10.1016/j.tetlet.2012.03.134
Myxobacterium-produced antibiotic TA (myxovirescin) inhibits type II signal peptidase
Xiao Y, Gerth K, Müller R, Wall D (2012)
Antimicrob. Agents Chemother. 56 (4): 2014-2021DOI: 10.1128/AAC.06148-11
Hyaladione, an S-methyl cyclohexadiene-dione from Hyalangium minutum
Okanya P, Mohr K, Gerth K, Steinmetz H, Huch V, Jansen R, Müller R (2012)
J. Nat. Prod. 75 (4): 768-770DOI: 10.1021/np200776v
Heterologous expression and genetic engineering of the tubulysin biosynthetic gene cluster using Red/ET recombineering and inactivation mutagenesis
Chai Y, Shan S, Weissman K, Hu S, Zhang Y, Müller R (2012)
Chem. Biol. 19 (3): 361-371DOI: 10.1016/j.chembiol.2012.01.007
Myxoprincomide: a natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome
Cortina N, Krug D, Plaza A, Revermann O, Müller R (2012)
Angew. Chem. Int. Ed. Engl. 51 (3): 811-816DOI: 10.1002/anie.201106305
Isolation and total synthesis of icumazoles and noricumazoles—antifungal antibiotics and cation-channel blockers from Sorangium cellulosum
Barbier J, Jansen R, Irschik H, Benson S, Gerth K, Böhlendorf B, Höfle G, Reichenbach H, Wegner J, Zeilinger C, Kirschning A, Müller R (2012)
Angew. Chem. Int. Ed. Engl. 51 (5): 1256-1260DOI: 10.1002/anie.201106435
Unusual carbon fixation gives rise to diverse polyketide extender units
Quade N, Huo L, Rachid S, Heinz D, Müller R (2012)
Nat. Chem. Biol. 8 (1): 117-124DOI: 10.1038/nchembio.734
Sandaracinus amylolyticus gen. nov., sp. nov., a starch-degrading soil myxobacterium, and description of Sandaracinaceae fam. nov
Mohr K, Garcia R, Gerth K, Irschik H, Müller R (2012)
Int. J. Syst. Evol. Microbiol. 62 (Pt 5): 1191-1198DOI: 10.1099/ijs.0.033696-0
Discovery of the rhizopodin biosynthetic gene cluster in Stigmatella aurantiaca Sg a15 by genome mining
Pistorius D, Müller R (2012)
Chembiochem : a European journal of chemical biology 13 (3): 416-426DOI: 10.1002/cbic.201100575
2011
Completing the puzzle of aurachin biosynthesis in Stigmatella aurantiaca Sg a15
Pistorius D, Li Y, Sandmann A, Müller R (2011)
Mol. Biosyst. 7 (12): 3308-3315DOI: 10.1039/c1mb05328k
AuaA, a membrane-bound farnesyltransferase from Stigmatella aurantiaca, catalyzes the prenylation of 2-methyl-4-hydroxyquinoline in the biosynthesis of aurachins
Stec E, Pistorius D, Müller R, Li S (2011)
ChemBioChem 12 (11): 1724-1730DOI: 10.1002/cbic.201100188
Unprecedented anthranilate priming involving two enzymes of the acyl adenylating superfamily in aurachin biosynthesis
Pistorius D, Li Y, Mann S, Müller R (2011)
J. Am. Chem. Soc. 133 (32): 12362-12365DOI: 10.1021/ja203653w
A highly conjugated dihydroxylated C28 steroid from a myxobacterium
Gawas D, Garcia R, Huch V, Müller R (2011)
Journal of natural products 74 (5): 1281-1283DOI: 10.1021/np100682c
Bendigoles D-F, bioactive sterols from the marine sponge-derived Actinomadura sp. SBMs009
Simmons L, Kaufmann K, Garcia R, Schwär G, Huch V, Müller R (2011)
Bioorganic & Medicinal Chemistry 19 (22): 6570-6575DOI: 10.1016/j.bmc.2011.05.044
Elansolid A3, a unique p-quinone methide antibiotic from Chitinophaga sancti
Jansen R, Gerth K, Steinmetz H, Reinecke S, Kessler W, Kirschning A, Müller R (2011)
Chem. Eur. J. 17 (28): 7739-7744DOI: 10.1002/chem.201100457
Molecular basis of elansolid biosynthesis: evidence for an unprecedented quinone methide initiated intramolecular Diels-Alder cycloaddition/macrolactonization
Dehn R, Katsuyama Y, Weber A, Gerth K, Jansen R, Steinmetz H, Höfle G, Müller R, Kirschning A (2011)
Angew. Chem. Int. Ed. Engl. 50 (17): 3882-3887DOI: 10.1002/anie.201006880
Roimatacene: an antibiotic against gram-negative bacteria isolated from Cystobacter ferrugineus Cb G35 (myxobacteria)
Zander W, Gerth K, Mohr K, Kessler W, Jansen R, Müller R (2011)
Chem. Eur. J. 17 (28): 7875-7881DOI: 10.1002/chem.201003677
p-Hydroxyacetophenone amides from Cystobacter ferrugineus, strain Cb G35
Zander W, Mohr K, Gerth K, Jansen R, Müller R (2011)
J. Nat. Prod. 74 (6): 1358-1363DOI: 10.1021/np1006789
Identification and characterization of the althiomycin biosynthetic gene cluster in Myxococcus xanthus DK897
Cortina N, Revermann O, Krug D, Müller R (2011)
ChemBioChem 12 (9): 1411-1416DOI: 10.1002/cbic.201100154
Fatty acid-related phylogeny of myxobacteria as an approach to discover polyunsaturated omega-3/6 fatty acids
Garcia R, Pistorius D, Stadler M, Müller R (2011)
J. Bacteriol. 193 (8): 1930-1942DOI: 10.1128/JB.01091-10
Biosynthesis of 2-alkyl-4(1H)-quinolones in Pseudomonas aeruginosa: potential for therapeutic interference with pathogenicity
Pistorius D, Ullrich A, Lucas S, Hartmann R, Kazmaier U, Müller R (2011)
ChemBioChem 12 (6): 850-853DOI: 10.1002/cbic.201100014
Marinoquinolines A-F, Pyrroloquinolines from Ohtaekwangia kribbensis (Bacteroidetes)
Okanya P, Mohr K, Gerth K, Jansen R, Müller R (2011)
J. Nat. Prod. 74 (4): 603-608DOI: 10.1021/np100625a
Insights into the complex biosynthesis of the leupyrrins in Sorangium cellulosum So ce690
Kopp M, Irschik H, Gemperlein K, Buntin K, Meiser P, Weissman K, Bode H, Müller R (2011)
Mol. Biosyst. 7 (5): 1549-1563DOI: 10.1039/c0mb00240b
Mining the cinnabaramide biosynthetic pathway to generate novel proteasome inhibitors
Rachid S, Huo L, Herrmann J, Stadler M, Köpcke B, Bitzer J, Müller R (2011)
ChemBioChem 12 (6): 922-931DOI: 10.1002/cbic.201100024
Elansolid A, a unique macrolide antibiotic from Chitinophaga sancti isolated as two stable atropisomers
Steinmetz H, Gerth K, Jansen R, Schläger N, Dehn R, Reinecke S, Kirschning A, Müller R (2011)
Angew. Chem. Int. Ed. Engl. 50 (2): 532-536DOI: 10.1002/anie.201005226
2010
Heterologous hosts
Zhang Y, Fu J, Bian X, Stewart A, Müller R (2010)
Patent (US 9,580,717 B2)
The CYPome of Sorangium cellulosum So ce56 and identification of CYP109D1 as a new fatty acid hydroxylase
Khatri Y, Hannemann F, Ewen K, Pistorius D, Perlova O, Kagawa N, Brachmann A, Müller R, Bernhardt R (2010)
Chem. Biol. 17: 1295-1305DOI: 10.1016/j.chembiol.2010.10.010
Production of omega-3 fatty acids by myxobacteria International Patent
Stadler M, Roemer E, Müller R, Garcia R, Pistorius D, Brachmann A (2010)
Patent (International Patent WO 2010/063451 A2)
Volatile Methyl Esters of Medium Chain Length from the Bacterium Chitinophaga Fx7914
Nawrath T, Gerth K, Müller R, Schulz S (2010)
Chemistry & Biodiversity 7 (9): 2228-2253
The Biosynthesis of the Aroma Volatile 2-Methyltetrahydrothiophen-3-one in the Bacterium Chitinophaga Fx7914
Nawrath T, Gerth K, Müller R, Schulz S (2010)
ChemBioChem 11 (13): 1914-1919DOI: 10.1002/cbic.201000296
Expanded phylogeny of myxobacteria and evidence for cultivation of the 'unculturables'
Garcia R, Gerth K, Stadler M, Dogma Jr. I, Müller R (2010)
Mol. Phylogenet. Evol. 57 (2): 878-887DOI: 10.1016/j.ympev.2010.08.028
Insights into an unusual nonribosomal peptide synthetase biosynthesis: identification and characterization of the GE81112 biosynthetic gene cluster
Binz T, Maffioli S, Sosio M, Donadio S, Müller R (2010)
J. Biol. Chem. 285 (43): 32710-32719DOI: 10.1074/jbc.M110.146803
Analysis of the sorangicin gene cluster reinforces the utility of a combined phylogenetic/retrobiosynthetic analysis for deciphering natural product assembly by trans-AT PKS
Irschik H, Kopp M, Weissman K, Buntin K, Piel J, Müller R (2010)
ChemBioChem 11 (13): 1840-1849DOI: 10.1002/cbic.201000313
Functions of genes and enzymes involved in phenalinolactone biosynthesis
Daum M, Schnell H, Herrmann S, Günther A, Murillo R, Müller R, Bisel P, Müller M, Bechtlold A (2010)
ChemBioChem 11 (10): 1383-1391DOI: 10.1002/cbic.201000117
Myxobacterial secondary metabolites: bioactivities and modes-of-action
Weissman K, Müller R (2010)
Nat. Prod. Rep. 27 (9): 1276-1295DOI: 10.1039/c001260m
Biosynthesis of thuggacins in myxobacteria: comparative cluster analysis reveals basis for natural product structural diversity
Buntin K, Irschik H, Weissman K, Luxenburger E, Blöcker H, Müller R (2010)
Chem. Biol. 17 (4): 342-356DOI: 10.1016/j.chembiol.2010.02.013
Biosynthesis of the myxobacterial antibiotic corallopyronin A
Erol Ö, Schäberle T, Schmitz A, Rachid S, Gurgui C, El Omari M, Lohr F, Kehraus S, Piel J, Müller R, König G (2010)
ChemBioChem 11 (9): 1235-1265DOI: 10.1002/cbic.201000085
Carolacton - a Macrolide Ketocarbonic Acid Reducing Biofilm Formation by the Caries- and Endocarditis-associated Bacterium Streptococcus mutans
Jansen R, Irschik H, Huch V, Schummer D, Steinmetz H, Bock M, Schmidt T, Kirschning A, Müller R (2010)
Eur. J. Org. Chem. 2010 (7): 1284-1289DOI: 10.1002/ejoc.200901126
An unusual thioesterase promotes isochromanone ring formation in ajudazol biosynthesis
Buntin K, Weissman K, Müller R (2010)
ChemBioChem 11 (8): 1137-1146DOI: 10.1002/cbic.200900712
Insights into multienzyme docking in hybrid PKS-NRPS megasynthetases revealed by heterologous expression and genetic engineering
Li Y, Weissman K, Müller R (2010)
ChemBioChem 11 (8): 1069-1075DOI: 10.1002/cbic.201000103
Discovery of 23 natural tubulysins from Angiococcus disciformis An d48 and Cystobacter SBCb004
Chai Y, Pistorius D, Ullrich A, Weissman K, Kazmaier U, Müller R (2010)
Chem. Biol. 17 (3): 296-309DOI: 10.1016/j.chembiol.2010.01.016
Characterization of a novel type of oxidative decarboxylase involved in the biosynthesis of the styryl moiety of chondrochloren from an acylated tyrosine
Rachid S, Revermann O, Dauth C, Müller R (2010)
J. Biol. Chem. 285 (17): 12482-12489DOI: 10.1074/jbc.M109.079707
2009
Pretubulysin, a potent and chemically accessible tubulysin precursor from Angiococcus disciformis
Ullrich A, Chai Y, Pistorius D, Elnakady Y, Herrmann J, Weissman K, Kazmaier U, Müller R (2009)
Angew. Chem. Int. Ed. Engl. 48 (24): 4422-4425DOI: 10.1002/anie.200900406
Progress in the chemistry of organic natural products
Altmann K, Höfle G, Müller R, Mulzer J, Prantz K (2009)
CollectedWorks
Synthesis and Biological Evaluation of Pretubulysin and Derivatives
Ullrich A, Herrmann J, Müller R, Kazmaier U (2009)
Eur. J. Org. Chem. 2009 (36): 6367-6378
Genome mining in Sorangium cellulosum So ce56 - identification and characterization of the homologous electron transfer proteins of a myxobacterial cytochrome P450
Ewen K, Hannemann F, Khatri Y, Perlova O, Kappl R, Krug D, Hüttermann J, Müller R, Bernhardt R (2009)
J. Biol. Chem. 284 (42): 28590-28598DOI: 10.1074/jbc.M109.021717
The impact of genomics on the exploitation of the myxobacterial secondary metabolome
Wenzel S, Müller R (2009)
Nat. Prod. Rep. 26 (11): 1385-1407DOI: 10.1039/b817073h
Non-modular polyketide synthases in myxobacteria
Li Y, Müller R (2009)
Phytochemistry 70 (15-16): 1850-1857
Myxobacteria--'microbial factories' for the production of bioactive secondary metabolites
Wenzel S, Müller R (2009)
Mol. Biosyst. 5 (6): 567-574DOI: 10.1039/b901287g
The biosynthetic potential of myxobacteria and their impact on drug discovery
Wenzel S, Müller R (2009)
Curr. Opin. Drug Discov. Devel. 12 (2): 220-230
Structure and action of the myxobacterial chondrochloren halogenase CndH: a new variant of FAD-dependent halogenases
Buedenbender S, Rachid S, Müller R, Schulz G (2009)
J. Mol. Biol. 385 (2): 520-530DOI: 10.1016/j.jmb.2008.10.057
Novel expression hosts for complex secondary metabolite megasynthetases: Production of myxochromide in the thermopilic isolate Corallococcus macrosporus GT-2
Perlova O, Gerth K, Kuhlmann S, Zhang Y, Müller R (2009)
Microb. Cell Fact. 8 (1)DOI: 10.1186/1475-2859-8-1
Identification of additional players in the alternative biosynthesis pathway to isovaleryl-CoA in the myxobacterium Myxococcus xanthus
Bode H, Ring M, Schwär G, Altmeyer M, Kegler C, Jose I, Singer M, Müller R (2009)
ChemBioChem 10 (1): 128-140DOI: 10.1002/cbic.200800219
Unusual chemistry in the biosynthesis of the antibiotic chondrochlorens
Rachid S, Scharfe M, Blöcker H, Weissman K, Müller R (2009)
Chem. Biol. 16 (1): 70-81DOI: 10.1016/j.chembiol.2008.11.005
Discovery of additional members of the tyrosine aminomutase enzyme family and the mutational analysis of CmdF
Krug D, Müller R (2009)
ChemBioChem 10 (4): 741-750DOI: 10.1002/cbic.200800748
Phaselicystis flava gen. nov., sp. nov., an arachidonic acid-containing soil myxobacterium, and the description of Phaselicystidaceae fam. nov
Garcia R, Reichenbach H, Ring M, Müller R (2009)
Int. J. Syst. Evol. Microbiol. 59 (6): 1524-1530DOI: 10.1099/ijs.0.003814-0
A brief tour of myxobacterial secondary metabolism
Weissman K, Müller R (2009)
Bioorganic & Medicinal Chemistry 17 (6): 2121-2136DOI: 10.1016/j.bmc.2008.11.025
2008
Epothilones - a fascinating family of microtubule stabilizing antitumor agents
Mulzer J, Altmann K, Hofle G, Müller R, Prantz K (2008)
Comptes Rendus Chimie 11 (11-12): 1336-1368DOI: 10.1016/j.crci.2008.02.005
Isolation and structure revision of the actin-binding macrolide rhizopodin from Myxococcus stipitatus (Myxobacteria)
Jansen R, Steinmetz H, Sasse F, Schubert W, Hagelueken G, Albrecht S, Müller R (2008)
Tetrahedron Lett. 49 (40): 5796-5799DOI: 10.1016/j.tetlet.2008.07.132
NtcA-A negative regulator of secondary metabolite biosynthesis in Sorangium cellulosum
Rachid S, Gerth K, Müller R (2008)
J. Biotechnol. 140 (1-2): 135-142DOI: 10.1016/j.jbiotec.2008.10.010
Epothilones - A fascinating family of microtubule stabilizing antitumor agents
Mulzer J, Altmann K, Höfle G, Müller R, Prantz K (2008)
C. R. Chimie 11
A type I/type III polyketide synthase hybrid biosynthetic pathway for the structurally unique ansa compound kendomycin
Wenzel S, Bode H, Kochems I, Müller R (2008)
ChemBioChem 9 (16): 2711-2721DOI: 10.1002/cbic.200800456
Crystal structure of a molecular assembly line
Weissman K, Müller R (2008)
Angew. Chem. Int. Ed. Engl. 47 (44): 8344-8346DOI: 10.1002/anie.200803293
Stereochemical determination and complex biosynthetic assembly of etnangien, a highly potent RNA polymerase inhibitor from the myxobacterium Sorangium cellulosum
Menche D, Arikan F, Perlova O, Horstmann N, Ahlbrecht W, Wenzel S, Jansen R, Irschik H, Müller R (2008)
J. Am. Chem. Soc. 130 (43): 14234-14243DOI: 10.1021/ja804194c
DKxanthene biosynthesis - understanding the basis for diversity-oriented synthesis in myxobacterial secondary metabolism
Meiser P, Weissman K, Bode H, Krug D, Dickschat J, Sandmann A, Müller R (2008)
Chem. Biol. 15 (8): 771-781DOI: 10.1016/j.chembiol.2008.06.005
Two functionally redundant Sfp-type 4'-phosphopantetheinyl transferases differentially activate biosynthetic pathways in Myxococcus xanthus
Meiser P, Müller R (2008)
ChemBioChem 9 (10): 1549-1553DOI: 10.1002/cbic.200800077
Stereochemical determination of thuggacins A-C, highly active antibiotics from the myxobacterium Sorangium cellulosum
Bock M, Buntin K, Müller R, Kirschning A (2008)
Angew. Chem. Int. Ed. Engl. 47 (12): 2308-2311DOI: 10.1002/anie.200704897
Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition
Fu J, Wenzel S, Perlova O, Wang J, Gross F, Tang Z, Yin Y, Stewart A, Müller R, Zhang Y (2008)
Nucleic Acids Res. 36 (17)DOI: 10.1093/nar/gkn499
A transposon-based strategy to scale up myxothiazol production in myxobacterial cell factories
Sandmann A, Frank B, Müller R (2008)
J. Biotechnol. 135 (3): 255-261DOI: 10.1016/j.jbiotec.2008.05.001
Efficient mining of myxobacterial metabolite profiles enabled by liquid chromatography-electrospray ionization-time-of-flight mass spectrometry and compound-based principal component analysis
Krug D, Zurek G, Schneider B, Garcia R, Müller R (2008)
Anal. Chim. Acta 624 (1): 97-106DOI: 10.1016/j.aca.2008.06.036
Myxochelin biosynthesis: direct evidence for two- and four-electron reduction of a carrier protein-bound thioester
Li Y, Weissman K, Müller R (2008)
J. Am. Chem. Soc. 130 (24): 7554-7555DOI: 10.1021/ja8025278
Heterologous expression and genetic engineering of the phenalinolactone biosynthetic gene cluster by using Red/ET recombineering
Binz T, Wenzel S, Schnell H, Bechthold A, Müller R (2008)
ChemBioChem 9 (3): 447-454DOI: 10.1002/cbic.200700549
Protein-protein interactions in multienzyme megasynthetases
Weissman K, Müller R (2008)
ChemBioChem 9 (6): 826-848DOI: 10.1002/cbic.200700751
Substrate specificity of the acyl transferase domains of EpoC from the epothilone polyketide synthase
Petkovic H, Sandmann A, Challis I, Hecht H, Silakowski B, Low L, Beeston N, Kuscer E, Garcia-Bernardo J, Leadlay P, …, Wilkinson B, Müller R (2008)
Org. Biomol. Chem. 6 (3): 500-506DOI: 10.1039/b714804f
Production of the antifungal isochromanone ajudazols A and B in Chondromyces crocatus Cm c5: biosynthetic machinery and cytochrome P450 modifications
Buntin K, Rachid S, Scharfe M, Blöcker H, Weissman K, Müller R (2008)
Angew. Chem. Int. Ed. Engl. 47 (24): 4595-4599DOI: 10.1002/anie.200705569
Discovering the Hidden Secondary Metabolome of Myxococcus xanthus: a Study of Intraspecific Diversity
Krug D, Zurek G, Revermann O, Vos M, Velicer G, Müller R (2008)
Appl. Environ. Microbiol. 74 (10): 3058-3068DOI: 10.1128/AEM.02863-07
2007
Complete genome sequence of the myxobacterium Sorangium cellulosum
Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer M, Bartels D, Bekel T, Beyer S, Bode E, …, Pühler A, Müller R (2007)
Nat. Biotechnol. 25 (11): 1281-1289DOI: 10.1038/nbt1354
From genetic diversity to metabolic unity: studies on the biosynthesis of aurafurones and aurafuron-like structures in myxobacteria and streptomycetes
Frank B, Wenzel S, Bode H, Scharfe M, Blöcker H, Müller R (2007)
J. Mol. Biol. 374 (1): 24-38DOI: 10.1016/j.jmb.2007.09.015
Mutasynthesis-derived myxalamids and origin of the isobutyryl-CoA starter unit of myxalamid B
Bode H, Meiser P, Klefisch T, Cortina N, Krug D, Göhring A, Schwär G, Mahmud T, Elnakady Y, Müller R (2007)
ChemBioChem 8 (17): 2139-2144DOI: 10.1002/cbic.200700401
Mutational analysis of the myxovirescin biosynthetic gene cluster reveals novel insights into the functional elaboration of polyketide backbones
Simunovic V, Müller R (2007)
ChemBioChem 8 (11): 1273-1280DOI: 10.1002/cbic.200700153
Biosynthesis of (R)-beta-tyrosine and its incorporation into the highly cytotoxic chondramides produced by Chondromyces crocatus
Rachid S, Krug D, Weissman K, Müller R (2007)
J. Biol. Chem. 282 (30): 21810-21817DOI: 10.1074/jbc.M703439200
Myxobacterial natural product assembly lines: fascinating examples of curious biochemistry
Wenzel S, Müller R (2007)
Nat. Prod. Rep. 24 (6): 1211-1224DOI: 10.1039/b706416k
Evidence for the mode of action of the highly cytotoxic Streptomyces polyketide kendomycin
Elnakady Y, Rohde M, Sasse F, Backes C, Keller A, Lenhof H, Weissman K, Müller R (2007)
ChemBioChem 8 (11): 1261-1272DOI: 10.1002/cbic.200700050
SorF, a glycosyltransferase with promiscuous donor substrate specificity in vitro
Kopp M, Rupprath C, Irschik H, Bechthold A, Elling L, Müller R (2007)
ChemBioChem 8 (7): 813-819DOI: 10.1002/cbic.200700024
Spiroketal polyketide formation in Sorangium: Identification and analysis of the biosynthetic gene cluster for the highly cytotoxic spirangienes
Frank B, Knauber J, Steinmetz H, Scharfe M, Blöcker H, Beyer S, Müller R (2007)
Chem. Biol. 14 (2): 221-233DOI: 10.1016/j.chembiol.2006.11.013
3-Hydroxy-3-methylglutaryl-CoA-like synthases direct the formation of methyl and ethyl side groups in the biosynthesis of the antibiotic myxovirescin A
Simunovic V, Müller R (2007)
ChemBioChem 8 (5): 497-500DOI: 10.1002/cbic.200700017
Deciphering regulatory mechanisms for secondary metabolite production in the myxobacterium Sorangium cellulosum So ce56
Rachid S, Gerth K, Kochems I, Müller R (2007)
Mol. Microbiol. 63 (6): 1783-1796DOI: 10.1111/j.1365-2958.2007.05627.x
Reversible sugar transfer by glycosyltransferases as atool for natural product (bio)synthesis
Bode H, Müller R (2007)
Angew. Chem. Int. Ed. Engl. 46 (13): 2147-2150DOI: 10.1002/anie.200604671
A type II polyketide synthase from the gram-negative bacterium Stigmatella aurantiaca is involved in aurachin alkaloid biosynthesis
Sandmann A, Dickschat J, Jenke-Kodama H, Kunze B, Dittmann E, Müller R (2007)
Angew. Chem. Int. Ed. Engl. 46 (15): 2712-2716DOI: 10.1002/anie.200603513
2006
Analysis of myxobacterial secondary metabolism goes molecular
Bode H, Müller R (2006)
J. Ind. Microbiol. Biotechnol. 33 (7): 577-588DOI: 10.1007/s10295-006-0082-7
The unique DKxanthene secondary metabolite family from the myxobacterium Myxococcus xanthus is required for developmental sporulation
Meiser P, Bode H, Müller R (2006)
Proc. Natl. Acad. Sci. U.S.A. 103 (50): 19128-19133DOI: 10.1073/pnas.0606039103
Exploiting microbial diversity and bacterial genomics to find novel antibiotics
Müller R (2006)
Int. J. Med. Microbiol. 296S3 (42): 138-139
Metabolic engineering of Pseudomonas putida for methylmalonyl-CoA biosynthesis to enable complex heterologous secondary metabolite formation
Gross F, Ring M, Perlova O, Fu J, Schneider S, Gerth K, Kuhlmann S, Stewart A, Zhang Y, Müller R (2006)
Chem. Biol. 13 (12): 1253-1264DOI: 10.1016/j.chembiol.2006.09.014
Metabolic physiology of Pseudomonas putida for heterologous production of myxochromide
Stephan S, Heinzle E, Wenzel S, Krug D, Müller R, Wittmann C (2006)
Process Biochem. 41 (2006): 2146-2152DOI: 10.1016/j.procbio.2006.06.022
On the biosynthetic origin of methoxymalonyl-acyl carrier protein, the substrate for incorporation of 'glycolate' units into ansamitocin and soraphen A
Wenzel S, Williamson R, Grünanger C, Xu J, Gerth K, Martinez R, Moss S, Carroll B, Grond S, Unkefer C, Müller R, Floss H (2006)
J. Am. Chem. Soc. 128 (44): 14325-14336DOI: 10.1021/ja064408t
Proteome analysis of Myxococcus xanthus by off-line two-dimensional chromatographic separation using monolithic poly-(styrene-divinylbenzene) columns combined with ion-trap tandem mass spectrometry
Schley C, Altmeyer M, Swart R, Müller R, Huber C (2006)
J. Proteome Res. 5 (10): 2760-2768DOI: 10.1021/pr0602489
Biochemical characterization of MelJ and MelK - myxobacterial enzymes that transform an amide into a methyl ester
Müller I, Müller R (2006)
FEBS J. 273 (16): 3768-3778DOI: 10.1111/j.1742-4658.2006.05385.x
3-Hydroxy-3-methylglutaryl-coenzyme A (CoA) synthase is involved in the biosynthesis of isovaleryl-CoA in the myxobacterium Myxococcus xanthus during fruiting body formation
Bode H, Ring M, Schwär G, Kroppenstedt R, Kaiser D, Müller R (2006)
J. Bacteriol. 188 (18): 6524-6528DOI: 10.1128/JB.00825-06
A unique mechanism for methyl ester formation via an amide intermediate found in myxobacteria
Müller I, Weinig S, Steinmetz H, Kunze B, Veluthoor S, Mahmud T, Müller R (2006)
ChemBioChem 7 (8): 1197-1205DOI: 10.1002/cbic.200600057
Reconstitution of myxothiazol biosynthetic gene cluster by Red/ET recombination and heterologous expression in Myxococcus xanthus
Perlova O, Fu J, Kuhlmann S, Krug D, Stewart F, Zhang Y, Müller R (2006)
Appl. Environ. Microbiol. 72 (12): 7485-7494DOI: 10.1128/AEM.01503-06
Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis
Berner M, Krug D, Bihlmaier C, Vente A, Müller R, Bechthold A (2006)
J. Bacteriol. 188 (7): 2666-2673DOI: 10.1128/JB.188.7.2666–2673.2006
Myxovirescin biosynthesis is directed by hybrid polyketide synthases/ nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl CoA synthases and trans-acting acyltransferases
Simunovic V, Zapp J, Rachid S, Krug D, Meiser P, Müller R (2006)
ChemBioChem 7 (8): 1206-1220DOI: 10.1002/cbic.200600075
Development of simple media which allow investigations into the global regulation of chivosazol biosynthesis with Sorangium cellulosum So ce56
Müller R, Gerth K (2006)
J. Biotechnol. 121 (2): 192-200DOI: 10.1016/j.jbiotec.2005.10.012
Molecular and biochemical studies of chondramide formation - highly cytotoxic natural products from Chondromyces crocatus Cm c5
Rachid S, Krug D, Kochems I, Kunze B, Scharfe M, Blöcker H, Zabriski M, Müller R (2006)
Chem. Biol. 14: 667-681DOI: 10.1016/j.chembiol.2006.06.002
Establishment of a real-time PCR protocol for expression studies of secondary metabolite biosynthetic gene clusters in the G/C-rich myxobacterium Sorangium cellulosum So ce56
Kegler C, Gerth K, Müller R (2006)
J. Biotechnol. 121 (2): 201-212DOI: 10.1016/j.jbiotec.2005.10.007
Nonribosomal Peptide Biosynthesis: Point Mutations and Module Skipping Lead to Chemical Diversity
Wenzel S, Meiser P, Binz T, Mahmud T, Müller R (2006)
Angew. Chem. Int. Ed. Engl. 45 (14): 2296-2301DOI: 10.1002/anie.200503737
Identification of StiR, the first regulator of secondary metabolite formation in the myxobacterium Cystobacter fuscus Cb f17.1
Rachid S, Sasse F, Beyer S, Müller R (2006)
J. Biotechnol. 121 (4): 429-441DOI: 10.1016/j.jbiotec.2005.08.014
Bacterial type III polyketide synthases: Phylogenetic analysis and potential for the production of novel secondary metabolites by heterologous expression in pseudomonads
Gross F, Luniak N, Perlova O, Gaitatzis N, Jenke-Kodama H, Gerth K, Gottschalk D, Dittmann E, Müller R (2006)
Arch. Microbiol. 185 (1): 28-38DOI: 10.1007/s00203-005-0059-3
Identification and analysis of the chivosazol biosynthetic gene cluster from the myxobacterial model strain Sorangium cellulosum So ce56
Perlova O, Gerth K, Hans A, Kaiser O, Müller R (2006)
J. Biotechnol. 121 (2): 174-191DOI: 10.1016/j.jbiotec.2005.10.011
2005
Moderately thermophilic myxobacteria: novel potential for the production of natural products isolation and characterization
Gerth K, Müller R (2005)
Environ. Microbiol. 7 (6): 874-880DOI: 10.1111/j.1462-2920.2005.00761.x
Heterologe Expression von komplexen myxobakteriellen Naturstoff-Biosynthesewegen in Pseudomonaden
Wenzel S, Müller R (2005)
Biospektrum 11 (5): 628-631
Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways
Wenzel S, Müller R (2005)
Curr. Opin. Biotechnol. 16 (6): 594-606DOI: 10.1016/j.copbio.2005.10.001
Evolutionary implications of bacterial polyketide synthases
Jenke-Kodama H, Sandmann A, Müller R, Dittmann E (2005)
Mol. Biol. Evol. 22 (10): 2027-2039DOI: 10.1093/molbev/msi193
The impact of bacterial genomics on natural product research
Bode H, Müller R (2005)
Angew. Chem. Int. Ed. Engl. 44 (42): 6828-6846DOI: 10.1002/anie.200501080
Production of the tubulin destabilizer disorazol in Sorangium cellulosum: biosynthetic machinery and regulatory genes
Kopp M, Irschik H, Pradella S, Müller R (2005)
ChemBioChem 6 (7): 1277-1286DOI: 10.1002/cbic.200400459
Formation of novel secondary metabolites by bacterial multimodular assembly lines: deviations from text book biosynthetic logic
Wenzel S, Müller R (2005)
Curr. Opin. Chem. Biol. 9: 447-458DOI: 10.1016/j.cbpa.2005.08.001
Aurafuron A and B, new bioactive polyketides from Stigmatella aurantiaca and Archangium gephyra (myxobacteria)
Kunze B, Reichenbach H, Müller R, Höfle G (2005)
J. Antibiot. 58 (4): 244-251
A novel type of geosmin biosynthesis in myxobacteria
Dickschat J, Bode H, Mahmud T, Müller R, Schulz S (2005)
J. Org. Chem. 70 (13): 5174-5182DOI: 10.1021/jo050449g
Novel insights into siderophore formation in myxobacteria
Gaitatzis N, Kunze B, Müller R (2005)
ChemBioChem 6 (2): 365-374DOI: 10.1002/cbic.200400206
Heterologous expression of a myxobacterial natural products assembly line in pseudomonads via red/ET recombineering
Wenzel S, Gross F, Zhang Y, Fu J, Stewart F, Müller R (2005)
Chem. Biol. 12 (3): 349-356DOI: 10.1016/j.chembiol.2004.12.012
Structure and Biosynthesis of Myxochromides S1-3 in Stigmatella aurantiaca: Evidence for An Iterative Bacterial Type I Polyketide Synthase and for Module Skipping in Nonribosomal Peptide Biosynthesis
Wenzel S, Kunze B, Höfle G, Silakowski B, Scharfe M, Blöcker H, Müller R (2005)
ChemBioChem 6 (2): 375-385DOI: 10.1002/cbic.200400282
Posttranslational modification of myxobacterial carrier protein domains in Pseudomonas sp. by an intrinsic phosphopantetheinyl transferase
Gross F, Gottschalk D, Müller R (2005)
Appl. Microbiol. Biotechnol. 68 (1): 66-74DOI: 10.1007/s00253-004-1836-7
Biosynthesis of iso-fatty acids in myxobacteria: iso-even fatty acids are derived by alpha-oxidation of iso-odd fatty acids
Bode H, Dickschat J, Kroppenstedt R, Schulz S, Müller R (2005)
J. Am. Chem. Soc. 127 (2): 532-533DOI: 10.1021/ja043570y
A novel biosynthetic pathway to isovaleryl-CoA in myxobacteria: The involvement of the mevalonate pathway
Mahmud T, Wenzel S, Wan E, Wen K, Bode H, Gaitatzis N, Müller R (2005)
ChemBioChem 6 (2): 322-330DOI: 10.1002/cbic.200400261
2004
Methods for heterologous expression of secondary metabolites
Wenzel S, Gross F, Zhang Y, Stewart F, Müller R (2004)
Patent
Identification and analysis of the core biosynthetic machinery of tubulysin, a potent cytotoxin with potential anticancer activity
Sandmann A, Sasse F, Müller R (2004)
Chem. Biol. 11 (8): 1071-1079DOI: 10.1016/j.chembiol.2004.05.014
Unusual biosynthesis of leupyrrins in the myxobacterium Sorangium cellulosum
Bode H, Wenzel S, Irschik H, Höfle G, Müller R (2004)
Angew. Chem. Int. Ed. Engl. 43 (32): 4163-4167DOI: 10.1002/anie.200454240
Biosynthesis of volatiles by the myxobacterium Myxococcus xanthus
Dickschat J, Wenzel S, Bode H, Müller R, Schulz S (2004)
ChemBioChem 5 (6): 778-787DOI: 10.1002/cbic.200300813
Don't classify polyketide synthases
Müller R (2004)
Chem. Biol. 11 (1): 4-6DOI: 10.1016/j.chembiol.2004.01.005
Critical variations of conjugational DNA transfer into secondary metabolite multiproducing Sorangium cellulosum strains So ce12 and So ce56: development of a mariner-based transposon mutagenesis system
Kopp M, Irschik H, Gross F, Perlova O, Sandmann A, Gerth K, Müller R (2004)
J. Biotechnol. 107 (1): 29-40DOI: 10.1016/j.jbiotec.2003.09.013
2003
Melithiazol biosynthesis: further insights into myxobacterial PKS/NRPS systems and evidence for a new subclass of methyl transferases
Weinig S, Hecht H, Mahmud T, Müller R (2003)
Chem. Biol. 10 (10): 939-952DOI: 10.1016/j.chembiol.2003.09.012
Markerless mutations in the myxothiazol biosynthetic gene cluster: A delicate megasynthetase with a superfluous nonribosomal peptide synthetase domain
Weinig S, Mahmud T, Müller R (2003)
Chem. Biol. 10 (10): 953-960DOI: 10.1016/j.chembiol.2003.09.013
Myxobacteria: proficient producers of novel natural products with various biological activities - past and future biotechnological aspects with the focus on the genus Sorangium
Gerth K, Pradella S, Perlova O, Beyer S, Müller R (2003)
J. Biotechnol. 106: 233-253DOI: 10.1016/j.jbiotec.2003.07.015
The leupyrrins: A structurally unique family of secondary metabolites from the myxobacterium Sorangium cellulosum
Bode H, Irschik H, Wenzel S, Reichenbach H, Müller R, Höfle G (2003)
J. Nat. Prod. 66 (9): 1203-1206DOI: 10.1021/np030109v
Possibility of bacterial recruitment of plant genes associated with the biosynthesis of secondary metabolites
Bode H, Müller R (2003)
Plant Physiol. 132 (3): 1153-1161DOI: 10.1104/pp.102.019760.
Steroid biosynthesis in prokaryotes: identification of myxobacterial steroids and cloning of the first bacterial 2,3(S)-oxidosqualene cyclase from the myxobacterium Stigmatella aurantiaca
Bode H, Zeggel B, Silakowski B, Wenzel S, Reichenbach H, Müller R (2003)
Mol. Microbiol. 47 (2): 471-481DOI: 10.1046/j.1365-2958.2003.03309.x
2002
A novel biosynthetic pathway providing precursors for fatty acid biosynthesis and secondary metabolite formation in myxobacteria
Mahmud T, Bode H, Silakowski B, Kroppenstedt R, Xu M, Nordhoff S, Höfle G, Müller R (2002)
J. Biol. Chem. 277 (36): 32768-74DOI: 10.1074/jbc.M205222200
The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase
Gaitatzis N, Silakowski B, Kunze B, Nordsiek G, Blöcker H, Höfle G, Müller R (2002)
J. Biol. Chem. 277 (15): 13082-13090DOI: 10.1074/jbc.M111738200
2001
In vitro reconstitution of the myxochelin biosynthetic machinery of Stigmatella aurantiaca Sg a15: Biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases
Gaitatzis N, Kunze B, Müller R (2001)
Proc. Natl. Acad. Sci. USA 98 (20): 11136-11141DOI: 10.1073/pnas.201167098
The mtaA gene of the myxothiazol biosynthetic gene cluster from Stigmatella aurantiaca DW4/3-1 encodes a phosphopantetheinyl transferase that activates polyketide synthases and polypeptide synthetases
Gaitatzis N, Hans A, Müller R, Beyer S (2001)
J. Biochem. (Tokyo) 129 (1): 119-124
Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: the myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sga15
Silakowski B, Nordsiek G, Kunze B, Blöcker H, Müller R (2001)
Chem. Biol. 8 (1): 59-69DOI: 10.1016/S1074-5521(00)00056-9
Multiple hybrid polyketide synthase/non-ribosomal peptide synthetase gene clusters in the myxobacterium Stigmatella aurantiaca
Silakowski B, Kunze B, Müller R (2001)
Gene 275 (2): 233-240
