![[Translate to German:] [Translate to German:]](/fileadmin/HIPS/__processed__/7/c/csm_marine_Bakterien_ac1ae5878f.jpg)
Antiinfektiva aus Mikrobiota
Prof. Dr. Christine Beemelmanns
Die Verbreitung von Antibiotika-resistente humanpathogene Bakterien stellt eine steigende Bedrohung für die menschliche Gesundheit dar, und die Entwicklung neuer Anti-infektiva und ein verbessertes Verständnis ihrer Funktion und Wirkungsweise ist dringend notwendig. Unsere Abteilung verfolgt eine Mikrobiota-basierte Strategie, um neue Wirkstoffe aus Mikroorganismen zu identifizieren.
Unsere Forschung
Die Gruppe von Prof. Christine Beemelmanns fokussiert sich auf die Identifizierung und funktionelle Analyse von neuartigen anti-infektiven Naturstoffen aus mikrobiellen Gemeinschaften. Ko-kultivierungsstudien sowie Zell-basierte Assays in Kombination mit chemisch-analytischen und molekularbiologische Methoden werden zur Evaluierung neuer mikrobieller Naturstoff-Produzenten verwendet. Zur Strukturaufklärung der sekretierten Naturstoffe wendet die Gruppe etablierte und innovative metabolomische, aktivitäts- als auch Genom-geleitete Methoden an. Basierend auf den isolierten neuartigen Naturstoffen erfolgt die funktionale Analyse und Evaluierung ihres Wirkspektrums.
Team-Mitglieder

Prof. Dr. Christine Beemelmanns
Gruppenleiterin

Annette Herkströter
Assistentin

Dr. Sebastian Götze
Wissenschaftler

Dr. Marie Dayras
Postdoc

Dr Tanya Lee Decker
Postdoc

Dr. Yaming Liu
Postdoc

Dr. Yen Thi Hai Lam
Postdoc

Chia-Chi Peng
Doktorandin

Lin Min
Doktorandin

Martinus de Kruijff
Doktorand

Sven Balluff
Doktorand

Rebecca Kochems
Technische Assistentin
Forschungsprojekte
Was bedeutet „Mikrobiota“?
Mikrobielle Gemeinschaften (Mikrobiota) setzen sich aus einer Vielzahl verschiedener Bakterien, Pilze und Vertreter ein- und wenigzellige Eukaryoten, sowie Viren, zusammen. Mikrobiota befinden sich unter anderem auf menschlichen, tierischen und pflanzlichen Gewebeoberflächen, wo sie essentielle Funktionen für den Wirt einnehmen können. Die Zusammensetzung der Mikrobiota korreliert in vielen Fällen mit ihrer Lokalisation und damit ihrer Funktion.
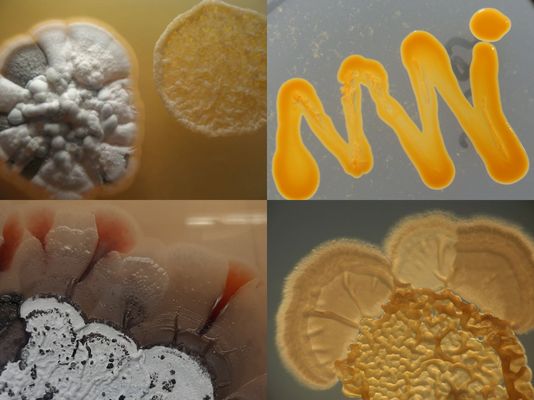
Mikroorganismen, die in der Abteilung MICA als Naturstoffproduzenten untersucht werden. ©Christine Beemelmanns
Welche Rolle spielen Naturstoffe?
Mikroorganismen regulieren und manipulieren ihr Zusammenleben durch die Aussendung von bioaktiven Naturstoffen. Mikrobielle Naturstoffe können antibiotisch wirken, um die Produzenten zu schützen, können aber auch als zelluläres Signal wirken, als Morphogen für den Wirtsorganismus oder als Nährstoff verstoffwechselt werden. Die chemischen Strukturen vieler dieser modulierenden Naturstoffe sind jedoch unbekannt, und damit ist ihr natürliche Funktion, sowie ihr Einfluss auf die Mikrobiota und möglichen Anwendungspotentiale bis heute nur sehr wenig erschlossen.
Wie kann das Naturstoffpotential erschlossen werden?
Da Naturstoffe wichtige Funktionen in mikrobiellen Interaktionen spielen, ist ihre Produktion eng mit der Zusammensetzung der Mikrobiota verknüpft. Die Beemelmanns-Gruppe analysiert repräsentative mikrobielle Gemeinschaften, um diesen chemischen Raum zu erschließen
Zielorientierte Bioassays basierend auf Bakterien-Bakterien und Bakterien-Pilz-Interaktionenstudien werden verwendet, um die Produktion von bioaktiven Naturstoffen zu stimulieren. Zur Strukturaufklärung der sekretierten Naturstoffe wendet die Gruppe etablierter und innovativer analytische Methoden an.
Die erhaltenen Naturstoffe werden auf ihre anti-infektive Wirkungsweise und Naturstoffe mit hohem Wirkpotential werde synthetisiert, um ihre Struktur-Wirkungsbeziehungen besser zu erfassen und die Profilierung der vielversprechendsten Kandidaten voranzutreiben. Zudem wird der Einfluss der charakterisierten Naturstoffe auf symbiotische Gemeinschaften mittels Mikrobiomstudien untersucht. Mittels dieser Studien erhoffen wir uns bessere Aussagen über die Stabilität und Dynamiken von mikrobiellen Gemeinschaften treffen zu können. Die Aufklärung der Biosynthesewege neuer Naturstoffe und ihre Regulation ist ebenfalls ein wichtiger Bestandteil unserer Forschung, um unser biokatalytisches Verständnis zu verbessern und die Entwicklung von biotechnologischen Ansätzen voranzutreiben.
Die Mitglieder der Abteilung haben unterschiedliche Wissenschaftliche Ausbildungs-Hintergründe. Hierzu zählen sowohl die synthetische organische Chemie, als auch Mikrobiologie, Biochemie, und Pharmazie.
Publikationen
2024
A type III polyketide synthase cluster in the phylum Planctomycetota is involved in alkylresorcinol biosynthesis
Milke L, Kabuu M, Zschoche R, Gätgens J, Krumbach K, Carlstedt K, Wurzbacher C, Balluff S, Beemelmanns C, Jogler C, Marienhagen J, Kallscheuer N (2024)
Appl Microbiol Biotechnol 108 (1)DOI: 10.1007/s00253-024-13065-x
Mining the microbiota for antibiotics
Beemelmanns C, Keller A, Müller R (2024)
Nat. Microbiol. 9 (1): 13-14DOI: 10.1038/s41564-023-01568-8
2023
Resolution of eleven reported and five novel Podaxis species based on ITS phylogeny, phylogenomics, morphology, ecology, and geographic distribution
Li G, Leal-Dutra C, Cuesta-Maté A, Conlon B, Peereboom N, Beemelmanns C, Aanen D, Rosendahl S, Debeer Z, Poulsen M (2023)
Persoonia (Molecular Phylogeny and Evolution of Fungi) 51: 257-279DOI: 10.3767/persoonia.2023.51.07
Heterologous expression of the cryptic mdk gene cluster and structural revision of maduralactomycin A
Schwitalla J, Le N, Um S, Schalk F, Brönstrup M, Baunach M, Beemelmanns C (2023)
RSC Adv 13 (48): 34136-34144DOI: 10.1039/D3RA05931F
Genome mining for macrolactam-encoding gene clusters allowed for the network-guided isolation of β-amino acid-containing cyclic derivatives and heterologous production of ciromicin A
Seibel E, Um S, Dayras M, Bodawatta K, Kruijff M, Jønsson K, Poulsen M, Kim K, Beemelmanns C (2023)
Commun Chem 6 (1): 1-15DOI: 10.1038/s42004-023-01034-w
Isolation of sulfonosphingolipids from the rosette-inducing bacterium Zobellia uliginosa and evaluation of their rosette-inducing activity
Peng C, Dormanns N, Regestein L, Beemelmanns C (2023)
RSC Adv 13 (39): 27520-27524DOI: 10.1039/d3ra04314b
Schatztruhe Menschliches Mikrobiom: Wie Naturstoffe Uns Beeinflus...: Ingenta Connect
Schumm C, Donate P, Hegemann J, Keller A, Beemelmanns C, Müller R (2023)
Pharmakon 11 (4): 290-297DOI: 10.1691/pn.20230032
Isolation, (bio)synthetic studies and evaluation of antimicrobial properties of drimenol-type sesquiterpenes of Termitomyces fungi
Kreuzenbeck N, Dhiman S, Roman D, Burkhardt I, Conlon B, Fricke J, Guo H, Blume J, Görls H, Poulsen M, …, Arndt H, Beemelmanns C (2023)
Commun Chem 6 (1)DOI: 10.1038/s42004-023-00871-z
Synthesis of Aryl‐ and Alkyl‐Containing 3‐Methylene‐5‐hydroxy Esters via a Barbier Allylation Reaction
Bartholomäus A, Roman D, Al-Jammal W, Vilotijevic I, Beemelmanns C (2023)
Eur. J. Org. Chem.DOI: 10.1002/ejoc.202300177
Adaptations of Pseudoxylaria towards a comb-associated lifestyle in fungus-farming termite colonies
Fricke J, Schalk F, Kreuzenbeck N, Seibel E, Hoffmann J, Dittmann G, Conlon B, Guo H, Wilhelm de Beer Z, Vassão D, …, Poulsen M, Beemelmanns C (2023)
ISME J.: 1-15DOI: 10.1038/s41396-023-01374-4
Multiple mutations in the Nav1.4 sodium channel of New Guinean toxic birds provide autoresistance to deadly batrachotoxin
Bodawatta K, Hu H, Schalk F, Daniel J, Maiah G, Koane B, Iova B, Beemelmanns C, Poulsen M, Jønsson K (2023)
Mol. Ecol.DOI: 10.1111/mec.16878
2022
The chemical ecology of the fungus-farming termite symbiosis
Schmidt S, Kildgaard S, Guo H, Beemelmanns C, Poulsen M (2022)
Nat. Prod. Rep. 39 (2): 231-248DOI: 10.1039/d1np00022e
A Modular Approach to the Antifungal Sphingofungin Family: Concise Total Synthesis of Sphingofungin A and C
Raguž L, Peng C, Kaiser M, Görls H, Beemelmanns C (2022)
Angewandte Chemie (International ed. in English) 61 (5)DOI: 10.1002/anie.202112616
Structural and Functional Analysis of Bacterial Sulfonosphingolipids and Rosette-Inducing Factor 2 (RIF-2) by Mass Spectrometry-Guided Isolation and Total Synthesis
Leichnitz D, Peng C, Raguž L, Rutaganira F, Jautzus T, Regestein L, King N, Beemelmanns C (2022)
Chem. Eur. J. 28 (8)DOI: 10.1002/chem.202103883
Signalling molecules inducing metamorphosis in marine organisms
Rischer M, Guo H, Beemelmanns C (2022)
Nat. Prod. Rep. 39 (9): 1833-1855DOI: 10.1039/d1np00073j
Application of pyrrolo-protected amino aldehydes in the stereoselective synthesis of anti-1,2-amino alcohols
Sauer M, Beemelmanns C (2022)
Chemical communications (Cambridge, England) 58 (64): 8990-8993DOI: 10.1039/d2cc02317b
Identification of the new prenyltransferase Ubi-297 from marine bacteria and elucidation of its substrate specificity
Amiri Moghaddam J, Guo H, Willing K, Wichard T, Beemelmanns C (2022)
Beilstein journal of organic chemistry 18: 722-731DOI: 10.3762/bjoc.18.72
GNPS-Guided Discovery of Madurastatin Siderophores from the Termite-Associated Actinomadura sp. RB99
Lee S, Schalk F, Schwitalla J, Guo H, Yu J, Song M, Jung W, Beer Z, Beemelmanns C, Kim K (2022)
Chem. Eur. J. 28 (36)DOI: 10.1002/chem.202200612
Comparative Genomic and Metabolomic Analysis of Termitomyces Species Provides Insights into the Terpenome of the Fungal Cultivar and the Characteristic Odor of the Fungus Garden of Macrotermes natalensis Termites
Kreuzenbeck N, Seibel E, Schwitalla J, Fricke J, Conlon B, Schmidt S, Hammerbacher A, Köllner T, Poulsen M, Hoffmeister D, Beemelmanns C (2022)
mSystems 7 (1)DOI: 10.1128/msystems.01214-21
Desaturation of the Sphingofungin Polyketide Tail Results in Increased Serine Palmitoyltransferase Inhibition
Hoefgen S, Bissell A, Huang Y, Gherlone F, Raguž L, Beemelmanns C, Valiante V (2022)
Microbiology Spectrum 10 (5)DOI: 10.1128/spectrum.01331-22
Insights into the Metabolomic Capacity of Podaxis and Isolation of Podaxisterols A-D, Ergosterol Derivatives Carrying Nitrosyl Cyanide-Derived Modifications
Guo H, Daniel J, Seibel E, Burkhardt I, Conlon B, Görls H, Vassão D, Dickschat J, Poulsen M, Beemelmanns C (2022)
Journal of natural products 85 (9): 2159-2167DOI: 10.1021/acs.jnatprod.2c00380
Synthesis of Functionalized δ-Hydroxy-β-keto Esters and Evaluation of Their Anti-inflammatory Properties
Grosse M, Günther K, Jordan P, Roman D, Werz O, Beemelmanns C (2022)
Chembiochem : a European journal of chemical biology 23 (9)DOI: 10.1002/cbic.202200073
Biosynthesis of the Sphingolipid Inhibitors Sphingofungins in Filamentous Fungi Requires Aminomalonate as a Metabolic Precursor
Bissell A, Rautschek J, Hoefgen S, Raguž L, Mattern D, Saeed N, Janevska S, Jojic K, Huang Y, Kufs J, …, Beemelmanns C, Valiante V (2022)
ACS chemical biology 17 (2): 386-394DOI: 10.1021/acschembio.1c00839
Total Synthesis and Functional Evaluation of IORs, Sulfonolipid-based Inhibitors of Cell Differentiation in Salpingoeca rosetta
Raguž L, Peng C, Rutaganira F, Krüger T, Stanišic A, Jautzus T, Kries H, Kniemeyer O, Brakhage A, King N, Beemelmanns C (2022)
Angewandte Chemie (International ed. in English)DOI: 10.1002/anie.202209105
2021
Species- and Caste-Specific Gut Metabolomes in Fungus-Farming Termites
Vidkjær N, Schmidt S, Hu H, Bodawatta K, Beemelmanns C, Poulsen M (2021)
Metabolites 11 (12)DOI: 10.3390/metabo11120839
Targeted Isolation of Saalfelduracin B-D from Amycolatopsis saalfeldensis Using LC-MS/MS-Based Molecular Networking
Um S, Seibel E, Schalk F, Balluff S, Beemelmanns C (2021)
Journal of natural products 84 (4): 1002-1011DOI: 10.1021/acs.jnatprod.0c01027
Comparative Genomic and Metabolic Analysis of Streptomyces sp. RB110 Morphotypes Illuminates Genomic Rearrangements and Formation of a New 46-Membered Antimicrobial Macrolide
Um S, Guo H, Thiengmag S, Benndorf R, Murphy R, Rischer M, Braga D, Poulsen M, Beer Z, Lackner G, Beemelmanns C (2021)
ACS chemical biology 16 (8): 1482-1492DOI: 10.1021/acschembio.1c00357
A community resource for paired genomic and metabolomic data mining
Schorn M, Verhoeven S, Ridder L, Huber F, Acharya D, Aksenov A, Aleti G, Moghaddam J, Aron A, Aziz S, …, Medema M, van der Hooft J (2021)
Nat Chem Biol 17 (4): 363-368DOI: 10.1038/s41589-020-00724-z
The Termite Fungal Cultivar Termitomyces Combines Diverse Enzymes and Oxidative Reactions for Plant Biomass Conversion
Schalk F, Gostincar C, Kreuzenbeck N, Conlon B, Sommerwerk E, Rabe P, Burkhardt I, Krüger T, Kniemeyer O, Brakhage A, …, Poulsen M, Beemelmanns C (2021)
mBio 12 (3)DOI: 10.1128/mBio.03551-20
GNPS-guided discovery of xylacremolide C and D, evaluation of their putative biosynthetic origin and bioactivity studies of xylacremolide A and B
Schalk F, Fricke J, Um S, Conlon B, Maus H, Jäger N, Heinzel T, Schirmeister T, Poulsen M, Beemelmanns C (2021)
RSC Adv 11 (31): 18748-18756DOI: 10.1039/d1ra00997d
Applications of the Horner–Wadsworth–Emmons Olefination in Modern Natural Product Synthesis
Roman D, Sauer M, Beemelmanns C (2021)
Synthesis 53 (16): 2713-2739DOI: 10.1055/a-1493-6331
Comparative Genomics Reveals Prophylactic and Catabolic Capabilities of Actinobacteria within the Fungus-Farming Termite Symbiosis
Murphy R, Benndorf R, Beer Z, Vollmers J, Kaster A, Beemelmanns C, Poulsen M (2021)
mSphere 6 (2)DOI: 10.1128/mSphere.01233-20
An integrative understanding of the large metabolic shifts induced by antibiotics in critical illness
Marfil-Sánchez A, Zhang L, Alonso-Pernas P, Mirhakkak M, Mueller M, Seelbinder B, Ni Y, Santhanam R, Busch A, Beemelmanns C, …, Bauer M, Panagiotou G (2021)
Gut microbes 13 (1)DOI: 10.1080/19490976.2021.1993598
DAnIEL: A User-Friendly Web Server for Fungal ITS Amplicon Sequencing Data
Loos D, Zhang L, Beemelmanns C, Kurzai O, Panagiotou G (2021)
Frontiers in microbiology 12DOI: 10.3389/fmicb.2021.720513
Two Distinct Bacterial Biofilm Components Trigger Metamorphosis in the Colonial Hydrozoan Hydractinia echinata
Guo H, Rischer M, Westermann M, Beemelmanns C (2021)
mBio 12 (3)DOI: 10.1128/mBio.00401-21
Revised structural assignment of azalomycins based on genomic and chemical analysis
Lee S, Guo H, Yu J, Park M, Dahse H, Jung W, Beemelmanns C, Kim K (2021)
Org. Chem. Front. 8 (17): 4791-4798DOI: 10.1039/D1QO00610J
Genome reduction and relaxed selection is associated with the transition to symbiosis in the basidiomycete genus Podaxis
Conlon B, Gostincar C, Fricke J, Kreuzenbeck N, Daniel J, Schlosser M, Peereboom N, Aanen D, Beer Z, Beemelmanns C, Gunde-Cimerman N, Poulsen M (2021)
iScience 24 (6)DOI: 10.1016/j.isci.2021.102680
Recent highlights of biosynthetic studies on marine natural products
Amiri Moghaddam J, Jautzus T, Alanjary M, Beemelmanns C (2021)
Org. Biomol. Chem. 19 (1): 123-140DOI: 10.1039/d0ob01677b
2020
Streptomyces smaragdinus sp. nov., isolated from the gut of the fungus growing-termite Macrotermes natalensis
Schwitalla J, Benndorf R, Martin K, Vollmers J, Kaster A, Beer Z, Poulsen M, Beemelmanns C (2020)
Int J Syst Evol Microbiol 70 (11): 5806-5811DOI: 10.1099/ijsem.0.004478
Targeted Discovery of Tetrapeptides and Cyclic Polyketide-Peptide Hybrids from a Fungal Antagonist of Farming Termites
Schalk F, Um S, Guo H, Kreuzenbeck N, Görls H, Beer Z, Beemelmanns C (2020)
Chembiochem : a European journal of chemical biology 21 (20): 2991-2996DOI: 10.1002/cbic.202000331
Modular Solid-Phase Synthesis of Antiprotozoal Barnesin Derivatives
Roman D, Raguž L, Keiff F, Meyer F, Barthels F, Schirmeister T, Kloss F, Beemelmanns C (2020)
Org. Lett. 22 (10): 3744-3748DOI: 10.1021/acs.orglett.0c00723
Polyhalogenation of Isoflavonoids by the Termite-Associated Actinomadura sp. RB99
Rak Lee S, Schalk F, Schwitalla J, Benndorf R, Vollmers J, Kaster A, Beer Z, Park M, Ahn M, Jung W, Beemelmanns C, Kim K (2020)
Journal of natural products 83 (10): 3102-3110DOI: 10.1021/acs.jnatprod.0c00676
Anti-adipogenic Pregnane Steroid from a Hydractinia-associated Fungus, Cladosporium sphaerospermum SW67
Lee S, Kang H, Yoo M, Yu J, Lee S, Yi S, Beemelmanns C, Lee J, Kim K (2020)
Natural Product Sciences 26 (3): 230-235DOI: 10.20307/nps.2020.26.3.230
Structure elucidation of the redox cofactor mycofactocin reveals oligo-glycosylation by MftF
Peña-Ortiz L, Graça A, Guo H, Braga D, Köllner T, Regestein L, Beemelmanns C, Lackner G (2020)
Chem. Sci. 11 (20): 5182-5190DOI: 10.1039/d0sc01172j
From Persian Gulf to Indonesia: interrelated phylogeographic distance and chemistry within the genus Peronia (Onchidiidae, Gastropoda, Mollusca)
Maniei F, Amiri Moghaddam J, Crüsemann M, Beemelmanns C, König G, Wägele H (2020)
Sci. Rep. 10 (1)DOI: 10.1038/s41598-020-69996-8
Absolute Configuration and Corrected NMR Assignment of 17-Hydroxycyclooctatin, a Fused 5-8-5 Tricyclic Diterpene
Lee S, Lee D, Park M, Lee J, Park H, Kang K, Kim C, Beemelmanns C, Kim K (2020)
Journal of natural products 83 (2): 354-361DOI: 10.1021/acs.jnatprod.9b00837
Xyloneside A: A New Glycosylated Incisterol Derivative from Xylaria sp. FB
Lee S, Kreuzenbeck N, Jang M, Oh T, Ko S, Ahn J, Beemelmanns C, Kim K (2020)
Chembiochem : a European journal of chemical biology 21 (16): 2253-2258DOI: 10.1002/cbic.202000065
Stereoselective Construction of ( E,Z )‐1,3‐Dienes and Its Application in Natural Product Synthesis
Hubert P, Seibel E, Beemelmanns C, Campagne J, Figueiredo R (2020)
Adv. Synth. Catal. 362 (24): 5532-5575DOI: 10.1002/adsc.202000730
Gene Cluster Activation in a Bacterial Symbiont Leads to Halogenated Angucyclic Maduralactomycins and Spirocyclic Actinospirols
Guo H, Schwitalla J, Benndorf R, Baunach M, Steinbeck C, Görls H, Beer Z, Regestein L, Beemelmanns C (2020)
Org. Lett. 22 (7): 2634-2638DOI: 10.1021/acs.orglett.0c00601
Immunomodulatory function of antimicrobial peptide EC-Hepcidin1 modulates the induction of inflammatory gene expression in primary cells of Caspian Trout (Salmo trutta caspius Kessler, 1877)
Ghodsi Z, Kalbassi M, Farzaneh P, Mobarez A, Beemelmanns C, Amiri Moghaddam J (2020)
Fish & shellfish immunology 104: 55-61DOI: 10.1016/j.fsi.2020.05.067
Nocardia macrotermitis sp. nov. and Nocardia aurantia sp. nov., isolated from the gut of the fungus-growing termite Macrotermes natalensis
Benndorf R, Schwitalla J, Martin K, Beer Z, Vollmers J, Kaster A, Poulsen M, Beemelmanns C (2020)
Int J Syst Evol Microbiol 70 (10): 5226-5234DOI: 10.1099/ijsem.0.004398
Actinomadura rubteroloni sp. nov. and Actinomadura macrotermitis sp. nov., isolated from the gut of the fungus growing-termite Macrotermes natalensis
Benndorf R, Martin K, Küfner M, Beer Z, Vollmers J, Kaster A, Beemelmanns C (2020)
Int J Syst Evol Microbiol 70 (10): 5255-5262DOI: 10.1099/ijsem.0.004403
2019
Fridamycin A, a Microbial Natural Product, Stimulates Glucose Uptake without Inducing Adipogenesis
Yoon S, Lee S, Hwang J, Benndorf R, Beemelmanns C, Chung S, Kim K (2019)
Nutrients 11 (4)DOI: 10.3390/nu11040765
Spirocyclic cladosporicin A and cladosporiumins I and J from a Hydractinia -associated Cladosporium sphaerospermum SW67
Rischer M, Lee S, Eom H, Park H, Vollmers J, Kaster A, Shin Y, Oh D, Kim K, Beemelmanns C (2019)
Org. Chem. Front. 6 (8): 1084-1093DOI: 10.1039/C8QO01104D
Ursprung und Funktionen der Sphingolipide
Raguz L, Beemelmanns C (2019)
Nachrichten aus der Chemie 67 (2): 66-70DOI: 10.1002/nadc.20194085913
Disease-free monoculture farming by fungus-growing termites
Otani S, Challinor V, Kreuzenbeck N, Kildgaard S, Krath Christensen S, Larsen L, Aanen D, Rasmussen S, Beemelmanns C, Poulsen M (2019)
Sci. Rep. 9 (1)DOI: 10.1038/s41598-019-45364-z
Stereoselective synthesis of unnatural (2S,3S)-6-hydroxy-4-sphingenine-containing sphingolipids
Leichnitz D, Pflanze S, Beemelmanns C (2019)
Org. Biomol. Chem. 17 (29): 6964-6969DOI: 10.1039/c9ob00990f
Hybrid Polyketides from a Hydractinia-Associated Cladosporium sphaerospermum SW67 and Their Putative Biosynthetic Origin
Lee S, Lee D, Eom H, Rischer M, Ko Y, Kang K, Kim C, Beemelmanns C, Kim K (2019)
Marine drugs 17 (11)DOI: 10.3390/md17110606
Beauvetetraones A–C, phomaligadione-derived polyketide dimers from the entomopathogenic fungus, Beauveria bassiana
Lee S, Küfner M, Park M, Jung W, Choi S, Beemelmanns C, Kim K (2019)
Org. Chem. Front. 6 (2): 162-166DOI: 10.1039/C8QO01009A
Efomycins K and L From a Termite-Associated Streptomyces sp. M56 and Their Putative Biosynthetic Origin
Klassen J, Lee S, Poulsen M, Beemelmanns C, Kim K (2019)
Frontiers in microbiology 10DOI: 10.3389/fmicb.2019.01739
Tropolone natural products
Guo H, Roman D, Beemelmanns C (2019)
Nat. Prod. Rep. 36 (8): 1137-1155DOI: 10.1039/c8np00078f
Reviewing the taxonomy of Podaxis: Opportunities for understanding extreme fungal lifestyles
Conlon B, Aanen D, Beemelmanns C, Beer Z, Fine Licht H, Gunde-Cimerman N, Schiøtt M, Poulsen M (2019)
Fungal Biology 123 (3): 183-187DOI: 10.1016/j.funbio.2019.01.001
Mechanistic characterization of three sesquiterpene synthases from the termite-associated fungus Termitomyces
Burkhardt I, Kreuzenbeck N, Beemelmanns C, Dickschat J (2019)
Org. Biomol. Chem. 17 (13): 3348-3355DOI: 10.1039/c8ob02744g
Metabolic Pathway Rerouting in Paraburkholderia rhizoxinica Evolved Long-Overlooked Derivatives of Coenzyme F420
Braga D, Last D, Hasan M, Guo H, Leichnitz D, Uzum Z, Richter I, Schalk F, Beemelmanns C, Hertweck C, Lackner G (2019)
ACS chemical biology 14 (9): 2088-2094DOI: 10.1021/acschembio.9b00605
Stereoselective Cascade Cyclizations with Samarium Diiodide to Tetracyclic Indolines: Precursors of Fluorostrychnines and Brucine
Beemelmanns C, Nitsch D, Bentz C, Reissig H (2019)
Chem. Eur. J. 25 (37): 8780-8789DOI: 10.1002/chem.201900087
2018
Role of Chemical Mediators in Aquatic Interactions across the Prokaryote-Eukaryote Boundary
Wichard T, Beemelmanns C (2018)
Journal of chemical ecology 44 (11): 1008-1021DOI: 10.1007/s10886-018-1004-7
Biosynthesis, Synthesis, and Activities of Barnesin A, a NRPS-PKS Hybrid Produced by an Anaerobic Epsilonproteobacterium
Rischer M, Raguž L, Guo H, Keiff F, Diekert G, Goris T, Beemelmanns C (2018)
ACS chemical biology 13 (8): 1990-1995DOI: 10.1021/acschembio.8b00445
The Inhibitory Effects of Cyclodepsipeptides from the Entomopathogenic Fungus Beauveria bassiana on Myofibroblast Differentiation in A549 Alveolar Epithelial Cells
Park Y, Lee S, Kim D, Yu J, Beemelmanns C, Chung K, Kim K (2018)
Molecules (Basel, Switzerland) 23 (10)DOI: 10.3390/molecules23102568
Chemical Identification of Isoflavonoids from a Termite-Associated Streptomyces sp. RB1 and Their Neuroprotective Effects in Murine Hippocampal HT22 Cell Line
Lee S, Song J, Song J, Ko H, Baek J, Trinh T, Beemelmanns C, Yamabe N, Kim K (2018)
International journal of molecular sciences 19 (9)DOI: 10.3390/ijms19092640
Natalenamides A⁻C, Cyclic Tripeptides from the Termite-Associated Actinomadura sp. RB99
Lee S, Lee D, Yu J, Benndorf R, Lee S, Lee D, Huh J, Beer Z, Kim Y, Beemelmanns C, Kang K, Kim K (2018)
Molecules (Basel, Switzerland) 23 (11)DOI: 10.3390/molecules23113003
Precursor-Directed Diversification of Cyclic Tetrapeptidic Pseudoxylallemycins
Guo H, Schmidt A, Stephan P, Raguž L, Braga D, Kaiser M, Dahse H, Weigel C, Lackner G, Beemelmanns C (2018)
Chembiochem : a European journal of chemical biology 19 (21): 2307-2311DOI: 10.1002/cbic.201800503
Expanding the Rubterolone Family: Intrinsic Reactivity and Directed Diversification of PKS-derived Pyrans
Guo H, Benndorf R, König S, Leichnitz D, Weigel C, Peschel G, Berthel P, Kaiser M, Steinbeck C, Werz O, Poulsen M, Beemelmanns C (2018)
Chem. Eur. J. 24 (44): 11319-11324DOI: 10.1002/chem.201802066
Natural Products from Actinobacteria Associated with Fungus-Growing Termites
Benndorf R, Guo H, Sommerwerk E, Weigel C, Garcia-Altares M, Martin K, Hu H, Küfner M, Beer Z, Poulsen M, Beemelmanns C (2018)
Antibiotics 7 (3)DOI: 10.3390/antibiotics7030083
2017
Linear Peptides Are the Major Products of a Biosynthetic Pathway That Encodes for Cyclic Depsipeptides
Wyche T, Ruzzini A, Beemelmanns C, Kim K, Klassen J, Cao S, Poulsen M, Bugni T, Currie C, Clardy J (2017)
Org. Lett. 19 (7): 1772-1775DOI: 10.1021/acs.orglett.7b00545
Bakterien-induzierte Morphogenese mariner Eukaryoten
Rischer M, Leichnitz D, Beemelmanns C (2017)
Biospektrum 23 (6): 634-637DOI: 10.1007/s12268-017-0848-7
Total synthesis and functional analysis of microbial signalling molecules
Leichnitz D, Raguž L, Beemelmanns C (2017)
Chem. Soc. Rev. 46 (20): 6330-6344DOI: 10.1039/c6cs00665e
Natural products and morphogenic activity of γ-Proteobacteria associated with the marine hydroid polyp Hydractinia echinata
Guo H, Rischer M, Sperfeld M, Weigel C, Menzel K, Clardy J, Beemelmanns C (2017)
Bioorg Med Chem 25 (22): 6088-6097DOI: 10.1016/j.bmc.2017.06.053
Isolation, Biosynthesis and Chemical Modifications of Rubterolones A-F: Rare Tropolone Alkaloids from Actinomadura sp. 5-2
Guo H, Benndorf R, Leichnitz D, Klassen J, Vollmers J, Görls H, Steinacker M, Weigel C, Dahse H, Kaster A, …, Poulsen M, Beemelmanns C (2017)
Chem. Eur. J. 23 (39): 9338-9345DOI: 10.1002/chem.201701005
Wie sich Bakterien schützen
Benndorf R, Leichnitz D, Rischer M, Beemelmanns C (2017)
Nachrichten aus der Chemie 65 (1): 21-25DOI: 10.1002/nadc.20174049020
Macrotermycins A-D, Glycosylated Macrolactams from a Termite-Associated Amycolatopsis sp. M39
Beemelmanns C, Ramadhar T, Kim K, Klassen J, Cao S, Wyche T, Hou Y, Poulsen M, Bugni T, Currie C, Clardy J (2017)
Org. Lett. 19 (5): 1000-1003DOI: 10.1021/acs.orglett.6b03831
2016
Bacterial lipids activate, synergize, and inhibit a developmental switch in choanoflagellates
Woznica A, Cantley A, Beemelmanns C, Freinkman E, Clardy J, King N (2016)
Proc. Natl. Acad. Sci. U.S.A. 113 (28): 7894-9DOI: 10.1073/pnas.1605015113
Draft Genome Sequence of Shewanella sp. Strain P1-14-1, a Bacterial Inducer of Settlement and Morphogenesis in Larvae of the Marine Hydroid Hydractinia echinata
Rischer M, Klassen J, Wolf T, Guo H, Shelest E, Clardy J, Beemelmanns C (2016)
Genome announc. 4 (1)DOI: 10.1128/genomeA.00003-16
A New Diketopiperazine, Cyclo(D-trans-Hyp-L-Leu) from a Kenyan Bacterium Bacillus licheniformis LB 8CT
Lee S, Beemelmanns C, Tsuma L, Clardy J, Cao S, Kim K (2016)
Natural product communications 11 (4): 461-3
Termisoflavones A-C, Isoflavonoid Glycosides from Termite-Associated Streptomyces sp. RB1
Kang H, Lee D, Benndorf R, Jung W, Beemelmanns C, Kang K, Kim K (2016)
Journal of natural products 79 (12): 3072-3078DOI: 10.1021/acs.jnatprod.6b00738
Pseudoxylallemycins A-F, Cyclic Tetrapeptides with Rare Allenyl Modifications Isolated from Pseudoxylaria sp. X802: A Competitor of Fungus-Growing Termite Cultivars
Guo H, Kreuzenbeck N, Otani S, Garcia-Altares M, Dahse H, Weigel C, Aanen D, Hertweck C, Poulsen M, Beemelmanns C (2016)
Org. Lett. 18 (14): 3338-41DOI: 10.1021/acs.orglett.6b01437
Isolation and Synthesis of a Bacterially Produced Inhibitor of Rosette Development in Choanoflagellates
Cantley A, Woznica A, Beemelmanns C, King N, Clardy J (2016)
J. Am. Chem. Soc. 138 (13): 4326-9DOI: 10.1021/jacs.6b01190
Natural products from microbes associated with insects
Beemelmanns C, Guo H, Rischer M, Poulsen M (2016)
Beilstein journal of organic chemistry 12: 314-27DOI: 10.3762/bjoc.12.34
2015
Draft Genome Sequences of Six Pseudoalteromonas Strains, P1-7a, P1-9, P1-13-1a, P1-16-1b, P1-25, and P1-26, Which Induce Larval Settlement and Metamorphosis in Hydractinia echinata
Klassen J, Wolf T, Rischer M, Guo H, Shelest E, Clardy J, Beemelmanns C (2015)
Genome announc. 3 (6)DOI: 10.1128/genomeA.01477-15
Genome Sequences of Three Pseudoalteromonas Strains (P1-8, P1-11, and P1-30), Isolated from the Marine Hydroid Hydractinia echinata
Klassen J, Rischer M, Wolf T, Guo H, Shelest E, Clardy J, Beemelmanns C (2015)
Genome announc. 3 (6)DOI: 10.1128/genomeA.01380-15
A new antibacterial octaketide and cytotoxic phenylethanoid glycosides from Pogostemon cablin (Blanco) Benth
Kim K, Beemelmanns C, Clardy J, Cao S (2015)
Bioorg Med Chem Lett 25 (14): 2834-6DOI: 10.1016/j.bmcl.2015.04.094
Strychnine as Target, Samarium Diiodide as Tool: A Personal Story
Beemelmanns C, Reissig H (2015)
Chemical record (New York, N.Y.) 15 (5): 872-85DOI: 10.1002/tcr.201500024
Evolution of a short route to strychnine by using the samarium-diiodide-induced cascade cyclization as a key step
Beemelmanns C, Reissig H (2015)
Chem. Eur. J. 21 (23): 8416-25DOI: 10.1002/chem.201500094
2014
Bacterial symbionts in agricultural systems provide a strategic source for antibiotic discovery
Ramadhar T, Beemelmanns C, Currie C, Clardy J (2014)
J. Antibiot. 67 (1): 53-8DOI: 10.1038/ja.2013.77
Natalamycin A, an Ansamycin from a Termite-Associated Streptomyces sp
Kim K, Ramadhar T, Beemelmanns C, Cao S, Poulsen M, Currie C, Clardy J (2014)
Chem. Sci. 5 (11): 4333-4338DOI: 10.1039/C4SC01136H
Naphthalenones and Isocoumarins from a Costa Rican Fungus Xylariaceae sp. CR1546C
Kim K, Beemelmanns C, Murillo C, Guillén A, Umaña L, Tamayo-Castillo G, Kim S, Clardy J, Cao S (2014)
Journal of Chemical Research 38 (12): 722-725DOI: 10.3184/174751914X14175406270662
Synthesis of the rosette-inducing factor RIF-1 and analogs
Beemelmanns C, Woznica A, Alegado R, Cantley A, King N, Clardy J (2014)
J. Am. Chem. Soc. 136 (29): 10210-3DOI: 10.1021/ja5046692
