Drug Design and Optimisation
Prof Dr Anna K. H. Hirsch
The Hirsch group adopts a target-based rational design strategy focusing on biologically relevant, often underexplored enzymes, transporters and regulators within bacterial and parasitic pathogens. The group employs a variety of biophysical methods to investigate compound–target interactions and has established several in vitro and cell-based assays for straightforward evaluation of novel anti-infectives and multiparameter optimisation.
Our research and approach
The emergence of drug-resistant strains of human pathogens such as P. aeruginosa, S. aureus or S. pneumoniae is a recognised threat to human health and urgently calls for the development of new antibacterial agents with novel modes of action. The department adopts a target-based strategy focussing on a portfolio of two types of drug targets.
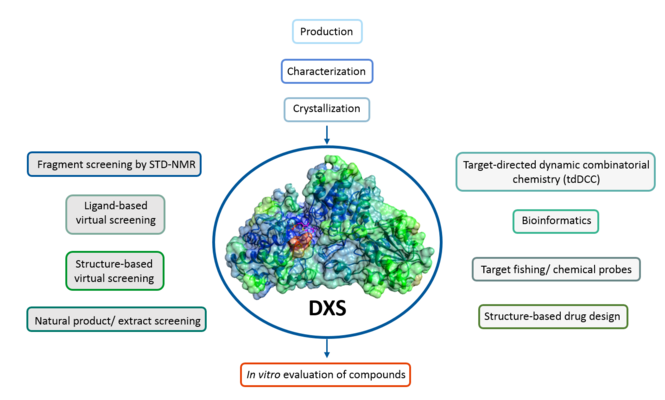
The first group comprises targets which impair vital mechanisms within the bacteria and effectively kill them. One example is the enzyme DXS which plays a crucial role in the methylerythritol phosphate pathway, which is essential for the biosynthesis of universal isoprenoid precursors in many gram-negative pathogens, but absent from humans. The second group comprises targets interfering with pathogenicity and virulence without affecting bacterial viability. These pathoblockers are believed to cause a lower rate of resistance development, whilst leaving the commensal microbiota untouched.
The Hirsch group applies a series of established hit-identification strategies, including structure- and fragment-based drug design, classical medicinal chemistry and virtual screening. In addition, pioneering of innovative protein-templated methods such as dynamic combinatorial chemistry and kinetic target-guided synthesis in terms of the scope of chemical reactions, biological targets and synergistic combinations addresses key bottlenecks. Use of established and innovative techniques to design, synthesize and profile the most promising inhibitors enables efficient subsequent multiparameter optimisation as well as elucidation of the mode of action.
Scientists in the interdisciplinary team have diverse backgrounds such as medicinal chemistry, synthetic organic chemistry, pharmacy, pharmacology, biology or biochemistry, resulting in a diverse skill set.
Team members

Prof Dr Anna K. H. Hirsch
Group Leader

Bahareh Kadkhodazadeh
Assistant

Nicole Klein Ramos
Assistant

Dr Godfrey Mayoka
Scientist

Dr Jörg Haupenthal
Scientist

Dr Andreas Kany
Postdoc

Dr Christian Schütz
Postdoc

Dr Danica Walsh
Postdoc

Dr Gwenaëlle Jézéquel
Postdoc

Dr Mariia Nesterkina
Postdoc

Dr Mostafa Hamed
Postdoc

Dr Walid A. M. Elgaher
Postdoc

Dr Života Selaković
Postdoc

Ahmed Amin
PhD Student

Andreas Klein
PhD Student

Angeliki Vgenopoulou
PhD Student

Antoine Lacour
PhD Student

Atanaz Shams
PhD Student

Daan Willocx
PhD Student

Dominik Kolling
PhD Student

Gabriele Bianchi
PhD Student

Hugo Scherer
PhD Student

Ioulia Antonia Exapicheidou
PhD Student

Justine Bassil
PhD Student

Márió Széles
PhD Student

Roya Shafiei
PhD Student

Sidra Eisa
PhD Student

Jannine Seelbach
Technical Assistant

Jeannine Jung
Technical Assistant

Lizana Martínez
Technical Assistant

Simone Amann
Technical Assistant

Tabea Wittmann
Technical Assistant
Research projects
Due to the rapidly increasing number of pathogenic bacteria being resistant to common antibiotics, the development of new antibacterial agents is urgently needed. To address these problems, the Hirsch group adopts a strategy based on the rational design and synthesis of anti-infectives with novel modes of action. In this context, the group is focusing on a portfolio of biologically relevant, ideally underexplored enzymes, transporters and regulators within bacterial pathogens. These drug targets can be grouped into those that impair vital mechanisms within the bacterium and effectively kill them (e.g., ECF-T, DnaN and DXS) and those that interfere with the pathogenicity and virulence without affecting bacterial viability (e.g., ColH and LasB).
The group employs a variety of biophysical methods to identify novel hit compounds and to investigate compound–target interactions. Various in vitro and cell-based assays, the generation of in silico data and the co-crystallization of selected compounds with their targets support the straightforward evaluation of novel anti-infectives and their further optimisation by classical and innovative medicinal chemistry approaches.
Below you will find an overview of selected targets we are currently addressing and to which we apply the above-mentioned strategies:
ECF-T
The energy-coupling factor (ECF) transporters are a family of transmembrane proteins involved in the uptake of vitamins by a wide range of bacteria. Inhibition of the activity of these proteins could reduce the viability of organisms that depend on vitamin uptake, including pathogenic species such as Staphylococcus aureus, Streptococcus pneumoniae and Enterococcus faecium. As these bacteria cannot synthesize important vitamins such as folic acid, pantothenate and niacin de novo, they depend on ECF transporters to take them up from the environment. Due to this central role in the metabolism of bacteria, ECF transporters are novel potential antimicrobial targets to tackle infection. Recently we identified the first classes of selective antibacterial agents showing promising inhibition of the ECF transporters. We are currently trying to further develop these compounds.
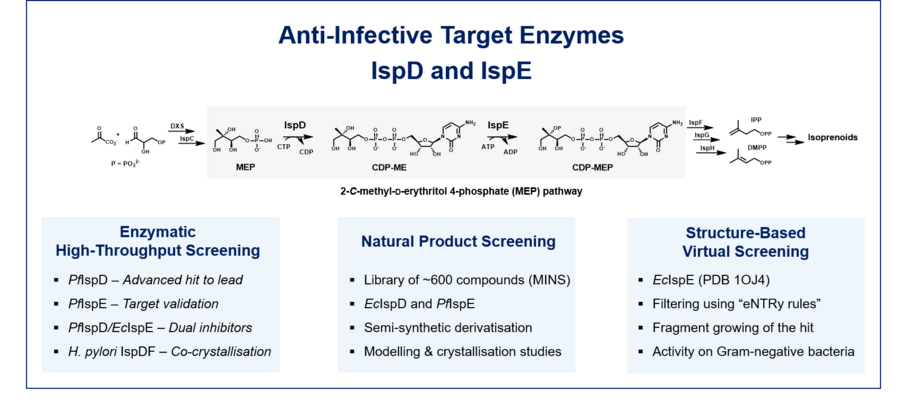
MEP pathway (target: DXS, IspD, IspE):
The methyl erythritol phosphate (MEP) pathway is a rich, underexplored source of anti-infective targets that are absent from humans but essential in many pathogenic bacteria, protozoa and plants. This opens up the possibility of developing highly selective inhibitors that are active against clinically relevant pathogens such as four ESKAPE pathogens: Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp., all of which have highest priority according to the WHO Global Priority List of antibiotic-resistant bacteria – as well as Mycobacterium tuberculosis and Plasmodium falciparum. In these pathogens, the MEP pathway is the sole source of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). The latter are the universal building blocks for the biosynthesis of isoprenoids, a large and diverse class of natural products fulfilling a myriad of indispensable functions. With DXS, IspD, IspE we address three essential enzymes of this pathway. Our best compounds are highly active against the mentioned pathogens and show nanomolar IC50 values.
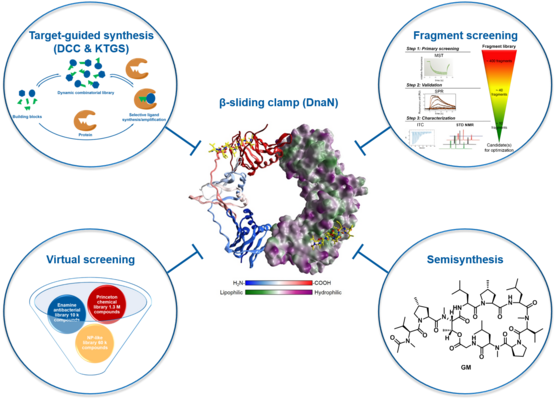
DnaN
Antibacterial drugs targeting essential multi-enzyme complexes are less prone to resistance development, since their malfunctioning is hard to bypass. In this context, the bacterial replisome machinery, which consists of at least twelve interacting enzymes that are highly conserved in bacteria, contains several attractive and promising antibacterial targets – including the bacterial β-sliding clamp DnaN. DnaN is a crucial subunit of the DNA polymerase III holoenzyme, preventing polymerase dissociation and enhancing polymerase activity. Furthermore, the β-sliding clamp shows interactions with a variety of other enzymes involved in replication and DNA repair processes. Its important functionality in the replication process and its highly conserved structure across bacterial species and its significant structural divergence to the mammalian counterpart (PCNA) make DnaN an attractive target for the treatment of various pathogens including Mycobacterium tuberculosis. Based on the natural product griselimycin (GM), a known DnaN binder, we adopted a semisynthetic approach, which resulted in a set of GM derivatives with promising antibacterial activities. Furthermore, we identified diverse small organic molecules binding to DnaN. The optimization of the most promising compounds is ongoing.
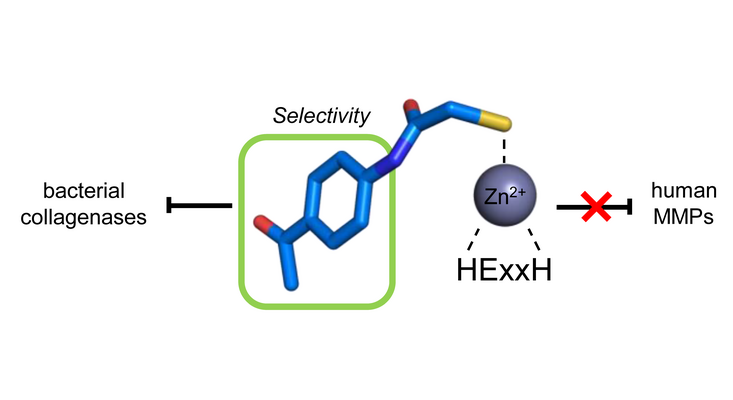
ColH
Clostridia represent a family of ubiquitously occurring gram-positive bacteria comprising perilous pathogens causing serious infectious diseases. The high lethality of these bacteria is related to collagenases, which are crucial for clostridial virulence, given their critical role in colonisation and evasion of host immune defence, acquisition of nutrients, facilitation of dissemination, or tissue damage during infection. The inhibition of these extracellular collagenases is conceptually attractive, as it does not attack the pathogen directly but rather blocks the colonisation and infiltration of the host by the clostridia. Targeting extracellular enzymes has the added benefit that inhibitors do not need to cross the bacterial cell wall, which has been challenging in many cases. Clostridial collagenases are zinc metalloproteinases with a multi-domain organisation, homologues of which are also found in many bacilli. Our group was the first who published compounds showing not only a highly potent inhibition of clostridial and bacillal collagenases (ColH IC50 in nM range) but also a strong selectivity over human MMPs.
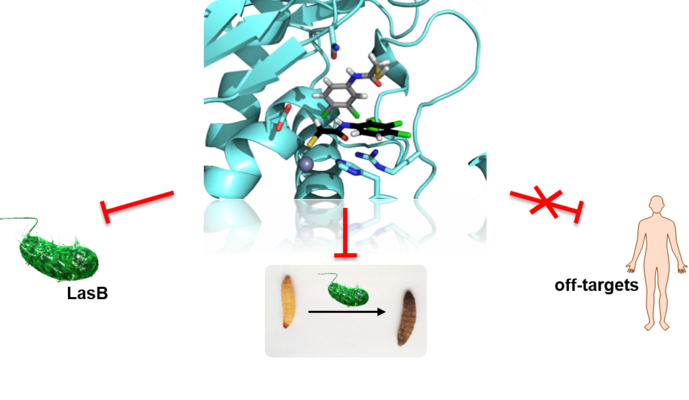
LasB
Pseudomonas aeruginosa (PA), a highly problematic gram-negative pathogen, which has been assigned critical priority by the WHO, is responsible for many nosocomial infections. The opportunistic pathogen is also commonly found in burns, and in lungs of COPD and cystic fibrosis patients. PA-derived virulence factors play pivotal roles in mechanisms mediating its pathogenesis and infectivity. The strategy to develop “pathoblockers” targeting such virulence factors is expected to leave the commensal microbiome intact and to be less prone to the development of resistance compared to conventional antibiotics. One of the major virulence factors of PA is the extracellular zinc-metalloprotease elastase LasB. LasB substantially contributes to disease progression in PA-infected individuals by facilitating host invasion and immune evasion, and was furthermore reported to be involved in the formation of PA biofilms. Inhibition of LasB is a promising strategy to effectively reduce virulence of PA without affecting viability of the pathogen and the host microbiome. Our most promising LasB inhibitors show nanomolar IC50 values and a promising safety profile.
Publications
2024
High Target Homology Does Not Guarantee Inhibition: Aminothiazoles Emerge as Inhibitors of Plasmodium falciparum
Johannsen S, Gierse R, Krüger A, Edwards R, Nanna V, Fontana A, Di Zhu, Masini T, Carvalho L, Poizat M, …, Wrenger C, Hirsch A (2024)
ACS Infect. Dis.DOI: 10.1021/acsinfecdis.3c00670
Hit optimization by dynamic combinatorial chemistry on Streptococcus pneumoniae energy-coupling factor transporter ECF-PanT
Exapicheidou I, Shams A, Ibrahim H, Tsarenko A, Backenköhler M, Hamed M, Diamanti E, Volkamer A, slotboom d, Hirsch A (2024)
Chem. Commun. 60 (7): 870-873DOI: 10.1039/D3CC04738E
2023
Clean Synthetic Strategies to Biologically Active Molecules from Lignin: A Green Path to Drug Discovery
Afanasenko A, Wu X, Santi A, Elgaher W, Kany A, Shafiei R, Schulze M, Schulz T, Haupenthal J, Hirsch A, Barta K (2023)
Angew. Chem. Int. Ed.DOI: 10.1002/anie.202308131
Identification of HuR–RNA Interfering Compounds by Dynamic Combinatorial Chemistry and Fluorescence Polarization
Della Volpe S, Listro R, Ambrosio F, Garbagnoli M, Linciano P, Rossi D, Costa G, Alcaro S, Vasile F, Hirsch A, Collina S (2023)
ACS Med. Chem. Lett.DOI: 10.1021/acsmedchemlett.3c00303
1-deoxy-D-xylulose-5-phosphate synthase from Pseudomonas aeruginosa and Klebsiella pneumoniae reveals conformational changes upon cofactor binding
Hamid R, Adam S, Lacour A, Monjas L, Köhnke J, Hirsch A (2023)
The Journal of biological chemistryDOI: 10.1016/j.jbc.2023.105152
pH-Responsive Dynaplexes as Potent Apoptosis Inductors by Intracellular Delivery of Survivin siRNA
Liu Y, Ashmawy S, Latta L, Weiss A, Kiefer A, Nasr S, Loretz B, Hirsch A, Lee S, Lehr C (2023)
Biomacromolecules 24 (8): 3742-3754DOI: 10.1021/acs.biomac.3c00424
Exploring the Translational Gap of a Novel Class of Escherichia coli IspE Inhibitors
Ropponen H, Diamanti E, Johannsen S, Illarionov B, Hamid R, Jaki M, Sass P, Fischer M, Haupenthal J, Hirsch A (2023)
ChemMedChemDOI: 10.1002/cmdc.202300346
Inhibitors of the Elastase LasB for the treatment of Pseudomonas aeruginosa lung infections
Konstantinovic J, Kany A, Alhayek A, Abdelsamie A, Sikandar A, Voos K, Yao Y, Andreas A, Shafiei R, Loretz B, …, Haupenthal J, Hirsch A (2023)
Artificial intelligence for natural product drug discovery
Mullowney M, Duncan K, Elsayed S, Garg N, van der Hooft J, Martin N, Meijer D, Terlouw B, Biermann F, Blin K, …, Robinson S, Medema M (2023)
Nat Rev Drug DiscovDOI: 10.1038/s41573-023-00774-7
Thermoresponsive cholesteric liquid-crystal systems doped with terpenoids as drug delivery systems for skin applications
Nesterkina M, Vashchenko O, Vashchenko P, Lisetski L, Kravchenko I, Hirsch A, Lehr C (2023)
European journal of pharmaceutics and biopharmaceutics : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.VDOI: 10.1016/j.ejpb.2023.09.002
Design of thiamine analogues for inhibition of thiamine diphosphate (ThDP)-dependent enzymes: Systematic investigation through Scaffold-Hopping and C2-Functionalisation
Chan A, Ho T, Irfan R, Hamid R, Rudge E, Iqbal A, Turner A, Hirsch A., Leeper F (2023)
Bioorganic chemistry 138DOI: 10.1016/j.bioorg.2023.106602
Facile Production of the Pseudomonas aeruginosa Virulence Factor LasB in E. coli for Structure-Based Drug Design
Kolling D, Haupenthal J, Hirsch A, Köhnke J (2023)
ChemBioChemDOI: 10.1002/cbic.202300185
Structure of Mycobacterium tuberculosis 1-Deoxy-D-Xylulose 5-Phosphate Synthase in Complex with Butylacetylphosphonate
Gawriljuk V, Oerlemans R, Gierse R, Jotwani R, Hirsch A, Groves M (2023)
Crystals 13 (5)DOI: 10.3390/cryst13050737
Not Every Hit-Identification Technique Works on 1-Deoxy-d-Xylulose 5-Phosphate Synthase (DXPS): Making the Most of a Virtual Screening Campaign
Johannsen S, Gierse R, Olshanova A, Smerznak E, Laggner C, Eschweiler L, Adeli Z, Hamid R, Alhayek A, Reiling N, Haupenthal J, Hirsch A (2023)
ChemMedChemDOI: 10.1002/cmdc.202200590
Genotoxic and mutational potential of monocyclic terpenoids (carvacrol, carvone and thymol) in Drosophila melanogaster
Nesterkina M, Bilokon S, Alieksieieva T, Kravchenko I, Hirsch A (2023)
Toxicol. Rep. 10DOI: 10.1016/j.toxrep.2023.02.009
Fighting antibiotic resistance-strategies and (pre)clinical developments to find new antibacterials
Walesch S, Birkelbach J, Jézéquel G, Haeckl F, Hegemann J, Hesterkamp T, Hirsch A, Hammann P, Müller R (2023)
EMBO reports 24 (1)DOI: 10.15252/embr.202256033
Towards Translation of PqsR Inverse Agonists: From In Vitro Efficacy Optimization to In Vivo Proof-of-Principle
Hamed M, Abdelsamie A, Rox K, Schütz C, Kany A, Röhrig T, Schmelz S, Blankenfeldt W, Arce-Rodriguez A, Borrero-de Acuña J, …, Hartmann R, Empting M (2023)
Adv. Sci. (Online)DOI: 10.1002/advs.202204443
2022
Discovery of the First Selective Nanomolar Inhibitors of ERAP2 by Kinetic Target‐Guided Synthesis
Camberlein V, Fléau C, Sierocki P, Li L, Gealageas R, Bosc D, Guillaume V, Warenghem S, Leroux F, Rosell M, …, Bouvier M, Deprez-Poulain R (2022)
Angew. Chemie 134 (39)DOI: 10.1002/ange.202203560
Synthesis, biological evaluation, and molecular docking studies of aldotetronic acid-based LpxC inhibitors
Wimmer S, Hoff K, Martin B, Grewer M, Denni L, Lascorz Massanet R, Raimondi M, Bülbül E, Melesina J, Hotop S, …, Sippl W, Holl R (2022)
Bioorganic chemistry 131DOI: 10.1016/j.bioorg.2022.106331
Proteoid biodynamers for safe mRNA transfection via pH-responsive nanorods enabling endosomal escape
Lee S, Nasr S, Rasheed S, Liu Y, Hartwig O, Kaya C, Boese A, Koch M, Herrmann J, Müller R, …, Hirsch A, Lehr C (2022)
Journal of controlled release : official journal of the Controlled Release Society 353: 915-929DOI: 10.1016/j.jconrel.2022.12.018
Baicalin lipid nanocapsules for treatment of glioma: characterization, mechanistic cytotoxicity, and pharmacokinetic evaluation
Ibrahim A, Abdel Gaber S, Fawzi Kabil M, Ahmed-Farid O, Hirsch A, El-Sherbiny I, Nasr M (2022)
Expert Opin. Drug Deliv. 19 (11): 1549-1560DOI: 10.1080/17425247.2022.2139370
Introduction to themed collection on fragment-based drug discovery
Rees D, Hirsch A, Erlanson D (2022)
RSC Med. Chem. 13DOI: 10.1039/D2MD90037H
An Efficient Way to Screen Inhibitors of Energy-Coupling Factor (ECF) Transporters in a Bacterial Uptake Assay
Bousis S, Winkler S, Haupenthal J, Fulco F, Diamanti E, Hirsch A (2022)
International journal of molecular sciences 23 (5)DOI: 10.3390/ijms23052637
Respiratory Syncytial Virus Two-Step Infection Screen Reveals Inhibitors of Early and Late Life Cycle Stages
Sake S, Kosch C, Blockus S, Haid S, Gunesch A, Zhang X, Friesland M, Trummer S, Grethe C, Kühnel A, …, Schulz T, Pietschmann T (2022)
Antimicrobial agents and chemotherapyDOI: 10.1128/aac.01032-22
Discovery and Characterization of Synthesized and FDA-Approved Inhibitors of Clostridial and Bacillary Collagenases
Alhayek A, Abdelsamie A, Schönauer E, Camberlein V, Hutterer E, Posselt G, Serwanja J, Blöchl C, Huber C, Haupenthal J, …, Wessler S, Hirsch A (2022)
Journal of medicinal chemistry 65 (19): 12933-12955DOI: 10.1021/acs.jmedchem.2c00785
Discovery of novel drug-like antitubercular hits targeting the MEP pathway enzyme DXPS by strategic application of ligand-based virtual screening
Di Zhu, Johannsen S, Masini T, Simonin C, Haupenthal J, Illarionov B, Andreas A, Awale M, Gierse R, van der Laan T, …, Reymond J, Hirsch A (2022)
Chem. Sci. 13 (36): 10686-10698DOI: 10.1039/D2SC02371G
Screening of Natural Products and Small Molecules Uncovers Novel Coronavirus 1a/1b Frameshifting Inhibitors with Antiviral Properties
Kibe A, Elgaher W, Rand U, Zimmer M, Kany A, Hermann J, Müller R, Cicin-Sain L, Hirsch A, Caliskan N (2022)
SSRN JournalDOI: 10.2139/ssrn.4157446
Design, Synthesis, Antimicrobial Activity, and Molecular Docking of Some New Diclofenac Derivatives
Tolba M, Hamed M, Sayed M, Kamal El-Dean A, Abdel-Mohsen S, Ibrahim O, Elgaher W, Hirsch A, Saddik A (2022)
Polycyclic Aromatic Compounds: 1-16DOI: 10.1080/10406638.2022.2102661
Structural analysis of 1-deoxy-D-xylulose 5-phosphate synthase from Pseudomonas aeruginosa and Klebsiella pneumoniae reveals conformational changes upon cofactor binding
Hamid R, Adam S, Lacour A, Gomez L, Hirsch A (2022)
bioRxivDOI: 10.1101/2022.07.04.498669
Identification of RAD51-BRCA2 Inhibitors Using N-Acylhydrazone-Based Dynamic Combinatorial Chemistry
Bagnolini G, Balboni B, Schipani F, Gioia D, Veronesi M, Franco F, Kaya C, Jumde R, Ortega J, Girotto S, …, Roberti M, Cavalli A (2022)
ACS Med. Chem. Lett. 13 (8): 1262-1269DOI: 10.1021/acsmedchemlett.2c00063
The Structures and Binding Modes of Small-Molecule Inhibitors of Pseudomonas aeruginosa Elastase LasB
Camberlein V, Jézéquel G, Haupenthal J, Hirsch A (2022)
Antibiotics 11 (8)DOI: 10.3390/antibiotics11081060
Transferring Microclusters of P. aeruginosa Biofilms to the Air-Liquid Interface of Bronchial Epithelial Cells for Repeated Deposition of Aerosolized Tobramycin
Horstmann J, Laric A, Boese A, Yildiz D, Röhrig T, Empting M, Frank N, Krug D, Müller R, Schneider-Daum N, Souza Carvalho-Wodarz C, Lehr C (2022)
ACS infectious diseases 8 (1): 137-149DOI: 10.1021/acsinfecdis.1c00444
Structure-Guided Optimization of Small-Molecule Folate Uptake Inhibitors Targeting the Energy-Coupling Factor Transporters
Kiefer A, Bousis S, Hamed M, Diamanti E, Haupenthal J, Hirsch A (2022)
J. Med. Chem.DOI: 10.1021/acs.jmedchem.1c02114
Citraconate inhibits ACOD1 (IRG1) catalysis, reduces interferon responses and oxidative stress, and modulates inflammation and cell metabolism
Chen F, Elgaher W, Winterhoff M, Büssow K, Waqas F, Graner E, Pires-Afonso Y, Casares Perez L, La Vega L, Sahini N, …, Hirsch A, Pessler F (2022)
Nature metabolism 4 (5): 534-546DOI: 10.1038/s42255-022-00577-x
Targeting Extracellular Bacterial Proteases for the Development of Novel Antivirulence Agents
Kaya C, Hirsch A (2022)
Chimia 76 (5)DOI: 10.2533/chimia.2022.402
Design and Synthesis of Novel Bis-Imidazolyl Phenyl Butadiyne Derivatives as HCV NS5A Inhibitors
Hamdy J, Emadeldin N, Hamed M, Frakolaki E, Katsamakas S, Vassilaki N, Zoidis G, Hirsch A, Abdel-Halim M, Abadi A (2022)
Pharmaceuticals 15 (5)DOI: 10.3390/ph15050632
First crystal structures of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Mycobacterium tuberculosis indicate a distinct mechanism of intermediate stabilization
Gierse R, Oerlemans R, Reddem E, Gawriljuk V, Alhayek A, Baitinger D, Jakobi H, Laber B, Lange G, Hirsch A, Groves M (2022)
Sci. Rep. 12 (1)DOI: 10.1038/s41598-022-11205-9
N-Aryl-2-iso-butylmercaptoacetamides: the discovery of highly potent and selective inhibitors of Pseudomonas aeruginosa virulence factor LasB and Clostridium histolyticum virulence factor ColH
Voos K, Yahiaoui S, Konstantinovic J, Schönauer E, Alhayek A, Sikandar A, Si Chaib K, Ramspoth T, Rox K, Haupenthal J, …, Ducho C, Hirsch A (2022)
ChemRxivDOI: 10.26434/chemrxiv-2022-fjrqr
Structure-Based Design of α-Substituted Mercaptoacetamides as Inhibitors of the Virulence Factor LasB from Pseudomonas aeruginosa
Kaya C, Walter I, Alhayek A, Shafiei R, Jézéquel G, Andreas A, Konstantinovic J, Schönauer E, Sikandar A, Haupenthal J, …, Hartmann R, Hirsch A (2022)
ACS Infect. Dis.DOI: 10.1021/acsinfecdis.1c00628
N-Aryl Mercaptopropionamides as Broad-Spectrum Inhibitors of Metallo-β-Lactamases
Kaya C, Konstantinovic J, Kany A, Andreas A, Kramer J, Brunst S, Weizel L, Rotter M, Frank D, Yahiaoui S, …, Wichelhaus T, Hirsch A (2022)
Journal of medicinal chemistry 65 (5): 3913-3922DOI: 10.1021/acs.jmedchem.1c01755
Metabolic profiling of S-praziquantel: Structure elucidation using the crystalline sponge method in combination with mass spectrometry and nuclear magnetic resonance
Rosenberger L, Jenniches J, Essen C, Khutia A, Kühn C, Marx A, Georgi K, Hirsch A, Hartmann R, Badolo L (2022)
Drug metabolism and disposition: the biological fate of chemicalsDOI: 10.1124/dmd.121.000663
Inhibition of Collagenase Q1 of Bacillus cereus as a Novel Antivirulence Strategy for the Treatment of Skin‐Wound Infections
Alhayek A, Khan E, Schönauer E, Däinghaus T, Shafiei R, Voos K, Han M, Ducho C, Posselt G, Wessler S, …, del Campo A, Hirsch A (2022)
Adv. Therap.DOI: 10.1002/adtp.202100222
Bacteriomimetic Liposomes Improve Antibiotic Activity of a Novel Energy-Coupling Factor Transporter Inhibitor
Drost M, Diamanti E, Fuhrmann K, Goes A, Shams A, Haupenthal J, Koch M, Hirsch A, Fuhrmann G (2022)
Pharmaceutics 14 (1)DOI: 10.3390/pharmaceutics14010004
Inhibitors of Pseudomonas aeruginosa virulence factor LasB
Ducho C, Hartmann R, Haupenthal J, Hirsch A, Kany A, Kaya C, Konstantinovic J, Voos K, Walter I, Yahiaoui S, …, Jumde R, Kiefer A (2022)
Patent A61K31/095; A61K31/10; A61K31/167; A61K31/4184; A61K31/426; A61K31/428; A61K31/4406; A61P31/04; C07C323/60; C07D213/74; C07D235/30; C07D277/46; C07D277/82; C07D333/36; C07F9/38; C07F9/40; (WO2022043322A1)
2021
Citatronic Acid and Derivates thereof for use as a medicament
Pessler F., Chen F., Winterhoff M., Büssow K., Blankenfeldt W., Hirsch A. K. H., Elgaher W (2021)
Patent (WO2022223778, 2022)
N-Phenyl-3-Mercaptopropanamide Derivatives as Metallo-Beta-Lactamase Inhibitors for the Treatment of Bacterial Infections
Hartmann R, Konstantinovic J, Haupenthal J, Hirsch A, Kany A, Kaya C, Yahiaoui S, Wichelhaus T, Proschak E (2021)
Patent A61K31/5375; A61P31/04; C07C233/15; C07C233/25; C07C233/33; C07C233/43; C07C233/54; C07C319/02; C07C323/52; C07D295/22; (WO2021191219A1)
New PqsR Inverse Agonist
Schütz C, Empting M, Ahmed S, Hamed M, Hartmann R, Röhrig T, Kany A, Hirsch A (2021)
Patent A61K31/4439; A61K31/506; A61P31/04; C07D401/12; C07D403/12; (WO2021136803A1)
Novel PqsR Inverse Agonists
Hamed M, Ahmed S, Empting. M., Schütz C, Hartmann R, Röhrig T, Kany A, Hirsch A (2021)
Patent A61K31/4439; A61P31/04; C07D401/14; C07D417/14; (WO2021136805A1)
Towards the sustainable discovery and development of new antibiotics
Miethke M, Pieroni M, Weber T, Brönstrup M, Hammann P, Halby L, Arimondo P, Glaser P, Aigle B, Bode H, …, Moser H, Müller R (2021)
Nature reviews. Chemistry 5 (10): 726-749DOI: 10.1038/s41570-021-00313-1
Targeting the IspD Enzyme in the MEP Pathway: Identification of a Novel Fragment Class
Diamanti E, Hamed M, Lacour A, Bravo P, Illarionov B, Fischer M, Rottmann M, Witschel M, Hirsch A (2021)
ChemMedChemDOI: 10.1002/cmdc.202100679
Substrate-Inspired Fragment Merging and Growing Affords Efficacious LasB Inhibitors
Kaya C, Walter I, Yahiaoui S, Sikandar A, Alhayek A, Konstantinovic J, Kany A, Haupenthal J, Köhnke J, Hartmann R, Hirsch A (2021)
Angewandte Chemie (International ed. in English)DOI: 10.1002/anie.202112295
Targeting the energy-coupling factor (ECF) transporters: identification of new tool compounds
Diamanti E, Setyawati I, Bousis S, Souza P, mojas l, Swier l, Haupenthal J, Gibson P, Volz C, stanek w, …, slotboom d, Hirsch A (2021)
ChemRxivDOI: 10.26434/chemrxiv-2021-xq08b-v2
Redesigning of the cap conformation and symmetry of the diphenylethyne core to yield highly potent pan-genotypic NS5A inhibitors with high potency and high resistance barrier
Abdallah M, Hamed M, Frakolaki E, Katsamakas S, Vassilaki N, Bartenschlager R, Zoidis G, Hirsch A, Abdel-Halim M, Abadi A (2021)
European journal of medicinal chemistryDOI: 10.1016/j.ejmech.2021.114034
Unveiling Adatoms in On-Surface Reactions: Combining Scanning Probe Microscopy with van't Hoff Plots
Moreno-López J, Pérez Paz A, Gottardi S, Solianyk L, Li J, Monjas L, Hirsch A, Mowbray D, Stöhr M (2021)
The journal of physical chemistry. C, Nanomaterials and interfaces 125 (18): 9847-9854DOI: 10.1021/acs.jpcc.1c03134
1st Spring Virtual Meeting on Medicinal Chemistry
Sousa M, Marques M, Faustino M (2021)
Chemistry Proceedings 4 (1)DOI: 10.3390/chemproc2021004001
Design, synthesis, and biological evaluation of novel benzimidazole derivatives as sphingosine kinase 1 inhibitor
Khairat S, Omar M, Ragab F, Roy S, Turab Naqvi A, Abdelsamie A, Hirsch A, Galal S, Hassan M, El Diwani H (2021)
Archiv der Pharmazie 354 (9)DOI: 10.1002/ardp.202100080
Identification of N,N-arylalkyl-picolinamide derivatives targeting the RNA-binding protein HuR, by combining biophysical fragment-screening and molecular hybridization
Della Volpe S, Linciano P, Listro R, Tumminelli E, Amadio M, Bonomo I, Elgaher W, Adam S, Hirsch A, Boeckler F, …, Rossi D, Collina S (2021)
Bioorganic chemistry 116DOI: 10.1016/j.bioorg.2021.105305
Expanding the Myxochelin Natural Product Family by Nicotinic Acid Containing Congeners
Frank N, Széles M, Akone S, Rasheed S, Hüttel S, Frewert S, Hamed M, Herrmann J, Schuler S, Hirsch A, Müller R (2021)
Molecules 26 (16)DOI: 10.3390/molecules26164929
N-Aryl mercaptoacetamides as potential multi-target inhibitors of metallo-β-lactamases (MBLs) and the virulence factor LasB from Pseudomonas aeruginosa
Yahiaoui S, Voos K, Haupenthal J, Wichelhaus T, Frank D, Weizel L, Rotter M, Brunst S, Kramer J, Proschak E, Ducho C, Hirsch A (2021)
RSC Med. Chem.DOI: 10.1039/D1MD00187F
Crystalline sponge affinity screening: A fast tool for soaking condition optimization without the need of X-ray diffraction analysis
Rosenberger L, Essen C, Khutia A, Kühn C, Georgi K, Hirsch A, Hartmann R, Badolo L (2021)
European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences 164DOI: 10.1016/j.ejps.2021.105884
A New PqsR Inverse Agonist Potentiates Tobramycin Efficacy to Eradicate Pseudomonas aeruginosa Biofilms
Schütz C, Ho D, Hamed M, Abdelsamie A, Röhrig T, Herr C, Kany A, Rox K, Schmelz S, Siebenbürger L, …, Lehr C, Empting M (2021)
Advanced science (Weinheim, Baden-Wurttemberg, Germany) 8 (12)DOI: 10.1002/advs.202004369
“Clicking“ fragment leads to novel dual-binding cholinesterase inhibitors
Moleda Z, Zawadzka A, Czarnocki Z, Monjas L, Hirsch A, Budzianowski A, Maurin J (2021)
Bioorg Med ChemDOI: 10.1016/j.bmc.2021.116269
Phosphonate as a Stable Zinc‐Binding Group for “Pathoblocker” Inhibitors of Clostridial Collagenase H (ColH) (ChemMedChem 8/2021)
Voos K, Schönauer E, Alhayek A, Haupenthal J, Andreas A, Müller R, Hartmann R, Brandstetter H, Hirsch A, Ducho C (2021)
ChemMedChem 16 (8): 1198-1198DOI: 10.1002/cmdc.202100229
Hit-optimization using target-directed dynamic combinatorial chemistry: Development of inhibitors of the anti-infective target 1-deoxy-D-xylulose-5-phosphate synthase
Jumde R, Guadigni M, Gierse R, Alhayek A, Di Zhu, Hamid Z, Johannsen S, Elgaher W, Neusens P, Nehls C, …, Reiling N, Hirsch A (2021)
Chemical ScienceDOI: 10.1039/D1SC00330E
Search for the Active Ingredients from a 2-Aminothiazole DMSO Stock Solution with Antimalarial Activity
Ropponen H, Bader C, Diamanti E, Illarionov B, Rottmann M, Fischer M, Witschel M, Müller R, Hirsch A (2021)
ChemMedChem 16 (13): 2089-2093DOI: 10.1002/cmdc.202100067
Mastering the Gram-Negative Bacterial Barrier - Chemical Approaches to Increase Bacterial Bioavailability of Antibiotics
Ropponen H, Richter R, Hirsch A, Lehr C (2021)
Advanced Drug Delivery ReviewsDOI: 10.1016/j.addr.2021.02.014
Assessment of the rules related to gaining activity against Gram-negative bacteria
Ropponen H, Diamanti E, Siemens A, Illarionov B, Haupenthal J, Fischer M, Rottmann M, Witschel M, Hirsch A (2021)
RSC Med. Chem.DOI: 10.1039/D0MD00409J
Enhancing glycan stability via site-selective fluorination: modulating substrate orientation by molecular design
Axer A, Jumde R, Adam S, Faust A, Schäfers M, Fobker M, Koehnke J, Hirsch A, Gilmour R (2021)
Chem. Sci. 12 (4): 1286-1294DOI: 10.1039/D0SC04297H
Identification of a 1-deoxy-D-xylulose-5-phosphate synthase (DXS) mutant with improved crystallographic properties
Gierse R, Reddem E, Alhayek A, Baitinger D, Hamid Z, Jakobi H, Laber B, Lange G, Hirsch A, Groves (2021)
Biochemical and Biophysical Research Communications 539DOI: 10.1016/j.bbrc.2020.12.069
2020
A hydrogel-based in vitro assay for the fast prediction of antibiotic accumulation in Gram-negative bacteria
Richter R, Kamal M, García-Rivera M, Kaspar J, Junk M, Elgaher W, Srikakulam S, Gress A, Beckmann A, Grißmer A, …, Schneider-Daum N, Lehr C (2020)
Materials today. Bio 8DOI: 10.1016/j.mtbio.2020.100084
Micro-rheological properties of lung homogenates correlate with infection severity in a mouse model of Pseudomonas aeruginosa lung infection
Murgia X, Kany A, Herr C, Ho D, Rossi C, Bals R, Lehr C, Hirsch A, Hartmann R, Empting M, Röhrig T (2020)
Scientific Reports 10 (1)DOI: 10.1038/s41598-020-73459-5
Semisynthesis and biological evaluation of amidochelocardin derivatives as broad-spectrum antibiotics
Grandclaudon C, Birudukota N, Elgaher W, Jumde R, Yahiaoui S, Arisetti N, Hennessen F, Hüttel S, Stadler M, Herrmann J, …, Hirsch A, Brönstrup M (2020)
European journal of medicinal chemistry 188DOI: 10.1016/j.ejmech.2019.112005
Discovery of Small-Molecule Stabilizers of 14-3-3 Protein-Protein Interactions via Dynamic Combinatorial Chemistry
Hartman A, Elgaher W, Hertrich N, Andrei S, Ottmann C, Hirsch A (2020)
ACS medicinal chemistry letters 11 (5): 1041-1046DOI: 10.1021/acsmedchemlett.9b00541
Evaluation of Bacterial RNA Polymerase Inhibitors in a Staphylococcus aureus-Based Wound Infection Model in SKH1 Mice
Haupenthal J, Kautz Y, Elgaher W, Pätzold L, Röhrig T, Laschke M, Tschernig T, Hirsch A, Molodtsov V, Murakami K, Hartmann R, Bischoff M (2020)
ACS infectious diseases 6 (10): 2573-2581DOI: 10.1021/acsinfecdis.0c00034
Protein‐Templated Hit Identification through an Ugi Four‐Component Reaction
Federica Mancini, M. Yagiz Unver, Walid A. M. Elgaher, Varsha R. Jumde, Alaa Alhayek, Peer Lukat, Jennifer Herrmann, Martin D. Witte, Matthias Köck, Wulf Blankenfeldt, Rolf Müller, Hirsch A (2020)
Chemistry – A European Journal 26 (64)DOI: 10.1002/chem.202086462
7-Hydroxycoumarins are Affinity-based Fluorescent Probes for Competitive Binding Studies of Macrophage Migration Inhibitory Factor
Xiao Z, Chen D, Song S, van der Vlag R, van der Wouden P, van Merkerk R, Cool R, Hirsch A, Melgert B, Quax W, Poelarends G, Dekker F (2020)
Journal of medicinal chemistryDOI: 10.1021/acs.jmedchem.0c01160
pH-Dependent morphology and optical properties of lysine-derived molecular biodynamers
Lee S, Kaya C, Jang H, Koch M, Loretz B, Buhler E, Lehr C, Hirsch A (2020)
Mater. Chem. Front. 4 (3): 905-909DOI: 10.1039/C9QM00651F
Rapid Discovery of Aspartyl Protease Inhibitors Using an Anchoring Approach
Konstantinidou M, Magari F, Sutanto F, Haupenthal J, Jumde R, Ünver M, Heine A, Camacho C, Hirsch A, Klebe G, Dömling A (2020)
ChemMedChem 15 (8): 680-684DOI: 10.1002/cmdc.202000024
Tracheal brush cells release acetylcholine in response to bitter tastants for paracrine and autocrine signaling
Hollenhorst M, Jurastow I, Nandigama R, Appenzeller S, Li L, Vogel J, Wiederhold S, Althaus M, Empting M, Altmüller J, …, Saliba A, Krasteva-Christ G (2020)
FASEB j. 34 (1): 316-332DOI: 10.1096/fj.201901314RR
Optimized Inhibitors of MDM2 via an Attempted Protein‐Templated Reductive Amination
Vlag R, Yagiz Unver M, Felicetti T, Twarda-Clapa A, Kassim F, Ermis C, Neochoritis C, Musielak B, Labuzek B, Dömling A, Holak T, Hirsch A (2020)
ChemMedChem 15 (4): 370-375DOI: 10.1002/cmdc.201900574
Synthesis and Biological Evaluation of Novel 2-Substituted Analogues of (–)-Pentenomycin I
Zisopoulou S, Bousis S, Haupenthal J, Herrmann J, Müller R, Hirsch A, Komiotis D, Gallos J, Stathakis C (2020)
Synlett 31 (05): 475-481DOI: 10.1055/s-0039-1690772
BOPC1 Enantiomers Preparation and HuR Interaction Study. From Molecular Modeling to a Curious DEEP-STD NMR Application
Della Volpe S, Listro R, Parafioriti M, Di Giacomo M, Rossi D, Ambrosio F, Costa G, Alcaro S, Ortuso F, Hirsch A, Vasile F, Collina S (2020)
ACS medicinal chemistry letters 11 (5): 883-888DOI: 10.1021/acsmedchemlett.9b00659
Validating the 1,2-Difluoro Motif As a Hybrid Bioisostere of CF3 and Et Using Matrix Metalloproteinases As Structural Probes
Erdeljac N, Thiehoff C, Jumde R, Daniliuc C, Höppner S, Faust A, Hirsch A, Gilmour R (2020)
Journal of medicinal chemistry 63 (11): 6225-6237DOI: 10.1021/acs.jmedchem.0c00648
Protein‐Templated Hit Identification via an Ugi Four‐Component Reaction
Mancini F, Unver M, Elgaher W, Jumde V, Alhayek A, Lukat P, Herrmann J, Witte M, Köck M, Blankenfeldt W, Müller R, Hirsch A (2020)
Chem. Eur. J.DOI: 10.1002/chem.202002250
N -Aryl-3-mercaptosuccinimides as Antivirulence Agents Targeting Pseudomonas aeruginosa Elastase and Clostridium Collagenases
Konstantinovic J, Yahiaoui S, Alhayek A, Haupenthal J, Schönauer E, Andreas A, Kany A, Müller R, Koehnke J, Berger F, …, Brandstetter H, Hirsch A (2020)
J. Med. Chem. 63 (15): 8359-8368DOI: 10.1021/acs.jmedchem.0c00584
Flotillin-mediated membrane fluidity controls peptidoglycan synthesis and MreB movement
Zielinska A, Savietto A, Sousa Borges A, Martinez D, Berbon M, Roelofsen J, Hartman A, Boer R, van der Klei I, Hirsch A, …, Bramkamp M, Scheffers D (2020)
eLife 9DOI: 10.7554/eLife.57179
Potential Dental Biofilm Inhibitors: Dynamic Combinatorial Chemistry Affords Sugar-Based Molecules that Target Bacterial Glucosyltransferase
Hartman A, Jumde V, Elgaher W, Te Poele E, Dijkhuizen L, Hirsch A (2020)
ChemMedChemDOI: 10.1002/cmdc.202000222
A rapid synthesis of low-nanomolar divalent LecA inhibitors in four linear steps from d -galactose pentaacetate
Zahorska E, Kuhaudomlarp S, Minervini S, Yousaf S, Lepsik M, Kinsinger T, Hirsch A, Imberty A, Titz A (2020)
Chem. Commun. 56 (62): 8822-8825DOI: 10.1039/d0cc03490h
Novel PqsR Inverse Agonists
Hamed M, Ahmed S, Empting. M., Schütz C, Hartmann R, Röhrig T, Kany A, Hirsch A (2020)
Patent (EP20150119)
2019
Energy-Coupling Factor Transporters as Novel Antimicrobial Targets
Bousis S, Setyawati I, Diamanti E, Slotboom D, Hirsch A (2019)
Advanced Therapeutics, 2, 1800066DOI: 10.1002/ADTP.201800066
Protein-Templated Dynamic Combinatorial Chemistry: Brief Overview and Experimental Protocol
Hartman A, Gierse R, Hirsch A (2019)
European journal of organic chemistry 2019 (22): 3581-3590DOI: 10.1002/ejoc.201900327
Concepts and Core Principles of Fragment-Based Drug Design
Kirsch P, Hartman A, Hirsch A, Empting M (2019)
Molecules (Basel, Switzerland) 24 (23)DOI: 10.3390/molecules24234309
Comparing the Self-Assembly of Sexiphenyl-Dicarbonitrile on Graphite and Graphene on Cu(111)
Schmidt N, Li J, Gottardi S, Moreno-Lopez J, Enache M, Monjas L, van der Vlag R, Havenith R, Hirsch A, Stöhr M (2019)
Chem. Eur. J. 25 (19): 5065-5070DOI: 10.1002/chem.201806312
Inverting Small Molecule–Protein Recognition by the Fluorine Gauche Effect: Selectivity Regulated by Multiple H→F Bioisosterism
Bentler P, Bergander K, Daniliuc C, Mück-Lichtenfeld C, Jumde R, Hirsch A, Gilmour R (2019)
Angewandte Chemie (International ed. in English) 58 (32): 10990-10994DOI: 10.1002/anie.201905452
Spray-drying of inhalable, multifunctional formulations for the treatment of biofilms formed in cystic fibrosis
Lababidi N, Ofosu Kissi E, Elgaher W, Sigal V, Haupenthal J, Schwarz B, Hirsch A, Rades T, Schneider M (2019)
Journal of Controlled Release 314: 62-71DOI: 10.1016/j.jconrel.2019.10.038
From Wood to Tetrahydro-2-benzazepines in Three Waste-Free Steps: Modular Synthesis of Biologically Active Lignin-Derived Scaffolds
Elangovan S, Afanasenko A, Haupenthal J, Sun Z, Liu Y, Hirsch A, Barta K (2019)
ACS Cent. Sci. 5 (10): 1707-1716DOI: 10.1021/acscentsci.9b00781
Surface state tunable energy and mass renormalization from homothetic quantum dot arrays
Piquero-Zulaica I, Li J, Abd El-Fattah Z, Solianyk L, Gallardo I, Monjas L, Hirsch A, Arnau A, Ortega J, Stöhr M, Lobo-Checa J (2019)
Nanoscale 11 (48): 23132-23138DOI: 10.1039/c9nr07365e
Novel Compounds Targeting the RNA-Binding Protein HuR. Structure-Based Design, Synthesis, and Interaction Studies
Della Volpe S, Nasti R, Queirolo M, Unver M, Jumde V, Dömling A, Vasile F, Potenza D, Ambrosio F, Costa G, …, Hirsch A, Collina S (2019)
ACS Med. Chem. Lett. 10 (4): 615-620DOI: 10.1021/acsmedchemlett.8b00600
A combinatorial approach for the discovery of drug-like inhibitors of 15-lipoxygenase-1
van der Vlag R, Guo H, Hapko U, Eleftheriadis N, Monjas L, Dekker F, Hirsch A (2019)
European journal of medicinal chemistry 174: 45-55DOI: 10.1016/j.ejmech.2019.04.021
Rational Adaptation of L3MBTL1 Inhibitors to Create Small-Molecule Cbx7 Antagonists
Simhadri C, Daze K, Douglas S, Milosevich N, Monjas L, Dev A, Brown T, Hirsch A, Wulff J, Hof F (2019)
ChemMedChem 14 (15): 1444-1456DOI: 10.1002/cmdc.201900021
Low-Dimensional Metal-Organic Coordination Structures on Graphene
Li J, Solianyk L, Schmidt N, Baker B, Gottardi S, Moreno Lopez J, Enache M, Monjas L, van der Vlag R, Havenith R, Hirsch A, Stöhr M (2019)
The journal of physical chemistry. C, Nanomaterials and interfaces 123 (20): 12730-12735DOI: 10.1021/acs.jpcc.9b00326
Novel 15-Lipoxygenase-1 Inhibitor Protects Macrophages from Lipopolysaccharide-Induced Cytotoxicity
Guo H, Verhoek I, Prins G, van der Vlag R, van der Wouden P, van Merkerk R, Quax W, Olinga P, Hirsch A, Dekker F (2019)
Journal of medicinal chemistry 62 (9): 4624-4637DOI: 10.1021/acs.jmedchem.9b00212
Replacement of an Indole Scaffold Targeting Human 15-Lipoxygenase-1 Using Combinatorial Chemistry
Prismawan D, van der Vlag R, Guo H, Dekker F, Hirsch A (2019)
HCA 102 (5)DOI: 10.1002/hlca.201900040
Reversible immobilization of a protein to a gold surface through multiple host-guest interactions
Schwarz D, Elgaher W, Hollemeyer K, Hirsch A, Wenz G (2019)
J. Mater. Chem. B 7 (40): 6148-6155DOI: 10.1039/c9tb00560a
2018
Trendbericht Biochemie 2017: Proteinvermittelte dynamische kombinatorische Chemie
Hirsch A (2018)
Nachr. Chem. 66 (3): 281-283DOI: 10.1002/nadc.20184071734
Druggability Assessment of Targets Used in Kinetic Target-Guided Synthesis
Unver M, Gierse R, Ritchie H, Hirsch A (2018)
Journal of medicinal chemistry 61 (21): 9395-9409DOI: 10.1021/acs.jmedchem.8b00266
Phage Display on the Anti-infective Target 1-Deoxy-d-xylulose-5-phosphate Synthase Leads to an Acceptor-Substrate Competitive Peptidic Inhibitor
Marcozzi A, Masini T, Di Zhu, Pesce D, Illarionov B, Fischer M, Herrmann A, Hirsch A (2018)
ChemBioChem 19 (1): 58-65DOI: 10.1002/cbic.201700402
Lipid-DNAs as Solubilizers of m THPC
Liu Y, de Vries J, Liu Q, Hartman A, Wieland G, Wieczorek S, Börner H, Wiehe A, Buhler E, Stuart M, …, Herrmann A, Hirsch A (2018)
Chem. Eur. J. 24 (4): 798-802DOI: 10.1002/chem.201705206
Glucansucrase (mutant) enzymes from Lactobacillus reuteri 180 efficiently transglucosylate Stevia component rebaudioside A, resulting in a superior taste
Te Poele E, Devlamynck T, Jäger M, Gerwig G, van de Walle D, Dewettinck K, Hirsch A, Kamerling J, Soetaert W, Dijkhuizen L (2018)
Sci Rep 8 (1)DOI: 10.1038/s41598-018-19622-5
Delivery system for budesonide based on lipid-DNA
Liu Y, Bos I, Oenema T, Meurs H, Maarsingh H, Hirsch A (2018)
European journal of pharmaceutics and biopharmaceutics : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V 130: 123-127DOI: 10.1016/j.ejpb.2018.06.012
Dynamic Proteoids Generated From Dipeptide-Based Monomers
Liu Y, Stuart M, Buhler E, Hirsch A (2018)
Macromolecular rapid communications 39 (13)DOI: 10.1002/marc.201800099
Design and Synthesis of Bioisosteres of Acylhydrazones as Stable Inhibitors of the Aspartic Protease Endothiapepsin
Jumde V, Mondal M, Gierse R, Unver M, Magari F, van Lier R, Heine A, Klebe G, Hirsch A (2018)
ChemMedChem 13 (21): 2266-2270DOI: 10.1002/cmdc.201800446
Exploration of ligand binding modes towards the identification of compounds targeting HuR: a combined STD-NMR and Molecular Modelling approach
Vasile F, Della Volpe S, Ambrosio F, Costa G, Unver M, Zucal C, Rossi D, Martino E, Provenzani A, Hirsch A, …, Potenza D, Collina S (2018)
Sci Rep 8 (1)DOI: 10.1038/s41598-018-32084-z
Donepezil-melatonin hybrids as butyrylcholinesterase inhibitors: Improving binding affinity through varying mode of linking fragments
Lozinska I, Swierczynska A, Moleda Z, Hartman A, Hirsch A, Czarnocki Z (2018)
Archiv der Pharmazie 351 (11)DOI: 10.1002/ardp.201800194
2017
Molecular Biodynamers: Dynamic Covalent Analogues of Biopolymers
Liu Y, Lehn J, Hirsch A (2017)
Accounts of chemical research 50 (2): 376-386DOI: 10.1021/acs.accounts.6b00594
DXS as a target for structure-based drug design
Gierse R, Redeem E, Diamanti E, Wrenger C, Groves M, Hirsch A (2017)
Future medicinal chemistry 9 (11): 1277-1294DOI: 10.4155/fmc-2016-0239
Molecular insight into specific 14-3-3 modulators: Inhibitors and stabilisers of protein-protein interactions of 14-3-3
Hartman A, Hirsch A (2017)
European journal of medicinal chemistry 136DOI: 10.1016/j.ejmech.2017.04.058
Compounds Interfering with Embryonic Lethal Abnormal Vision (ELAV) Protein-RNA Complexes: An Avenue for Discovering New Drugs
Nasti R, Rossi D, Amadio M, Pascale A, Unver M, Hirsch A, Collina S (2017)
Journal of medicinal chemistry 60 (20): 8257-8267DOI: 10.1021/acs.jmedchem.6b01871
Fine-tuning Nanocarriers Specifically toward Cargo: A Competitive Study on Solubilizing Related Photosensitizers for Photodynamic Therapy
Wieczorek S, Remmler D, Masini T, Kochovski Z, Hirsch A, Börner H (2017)
Bioconjugate chemistry 28 (3): 760-767DOI: 10.1021/acs.bioconjchem.6b00549
Pentapeptide‐rich peptidoglycan at the Bacillus subtilis cell‐division site
Morales Angeles D, Liu Y, Hartman A, Borisova M, Sousa Borges A, Kok N, Beilharz K, veening j, Mayer C, Hirsch A, Scheffers D (2017)
Molecular microbiology 104 (2): 319-333DOI: 10.1111/mmi.13629
Bicyclic enol cyclocarbamates inhibit penicillin-binding proteins
Dockerty P, Edens J, Tol M, Morales Angeles D, Domenech A, Liu Y, Hirsch A, veening j, Scheffers D, Witte M (2017)
Organic & biomolecular chemistry 15 (4): 894-910DOI: 10.1039/c6ob01664b
Designed Spiroketal Protein Modulation
Scheepstra M, Andrei S, Unver M, Hirsch A, Leysen S, Ottmann C, Brunsveld L, Milroy L (2017)
Angewandte Chemie (International ed. in English) 56 (20): 5480-5484DOI: 10.1002/anie.201612504
Insight into the complete substrate-binding pocket of ThiT by chemical and genetic mutations
Swier L, Monjas L, Reeßing F, Oudshoorn R, Aisyah, Primke T, Bakker M, van Olst E, Ritschel T, Faustino I, …, Hirsch A, Slotboom D (2017)
MedChemComm 8 (5): 1121-1130DOI: 10.1039/c7md00079k
Saccharide-Containing Dynamic Proteoids
Liu Y, Stuart M, Witte M, Buhler E, Hirsch A (2017)
Chemistry (Weinheim an der Bergstrasse, Germany) 23 (64): 16162-16166DOI: 10.1002/chem.201703584
Dynamic Combinatorial Chemistry to Identify Binders of ThiT, an S-Component of the Energy-Coupling Factor Transporter for Thiamine
Monjas L, Swier L, Setyawati I, slotboom d, Hirsch A (2017)
ChemMedChem 12 (20): 1693-1696DOI: 10.1002/cmdc.201700440
2016
Design and synthesis of thiamine analogues to study their binding to the ECF transporter for thiamine in bacteria
Monjas L, Swier L, Voogd A, Oudshoorn R, Hirsch A, Slotboom D (2016)
Med. Chem. Commun. 7 (5): 966-971DOI: 10.1039/C6MD00022C
Furoates and thenoates inhibit pyruvate dehydrogenase kinase 2 allosterically by binding to its pyruvate regulatory site
Masini T, Birkaya B, van Dijk S, Mondal M, Hekelaar J, Jäger M, van Terwisscha Scheltinga A, Patel M, Hirsch A, Moman E (2016)
Journal of enzyme inhibition and medicinal chemistry 31 (sup4): 170-175DOI: 10.1080/14756366.2016.1201812
Proteoid Dynamers with Tunable Properties
Liu Y, Stuart M, Buhler E, Lehn J, Hirsch A (2016)
Fragment Linking and Optimization of Inhibitors of the Aspartic Protease Endothiapepsin: Fragment-Based Drug Design Facilitated by Dynamic Combinatorial Chemistry
Mondal M, Radeva N, Fanlo-Virgós H, Otto S, Klebe G, Hirsch A (2016)
Angewandte Chemie (International ed. in English) 55 (32): 9422-6DOI: 10.1002/anie.201603074
Fragment-Based Drug Design Facilitated by Protein-Templated Click Chemistry: Fragment Linking and Optimization of Inhibitors of the Aspartic Protease Endothiapepsin
Mondal M, Unver M, Pal A, Bakker M, Berrier S, Hirsch A (2016)
Chem. Eur. J. 22 (42): 14826-14830DOI: 10.1002/chem.201603001
2015
Dynamic combinatorial chemistry: a tool to facilitate the identification of inhibitors for protein targets
Mondal M, Hirsch A (2015)
Chemical Society reviews 44 (8): 2455-88DOI: 10.1039/c4cs00493k
Fighting malaria: structure-guided discovery of nonpeptidomimetic plasmepsin inhibitors
Huizing A, Mondal M, Hirsch A (2015)
Journal of medicinal chemistry 58 (13): 5151-63DOI: 10.1021/jm5014133
Harnessing dynamic combinatorial chemistry in the search for new ligands for protein targets
Monjas L, Hirsch A (2015)
Future medicinal chemistry 7 (16): 2095-8DOI: 10.4155/fmc.15.146
Combinatorial screening for specific drug solubilizers with switchable release profiles
Wieczorek S, Vigne S, Masini T, Ponader D, Hartmann L, Hirsch A, Börner H (2015)
Macromolecular Bioscience 15 (1): 82-9DOI: 10.1002/mabi.201400443
Structure-based design of potent small-molecule binders to the S-component of the ECF transporter for thiamine
Swier L, Monjas L, Guskov A, Voogd A, Erkens G, slotboom d, Hirsch A (2015)
ChemBioChem 16 (5): 819-26DOI: 10.1002/cbic.201402673
Fragment growing exploiting dynamic combinatorial chemistry of inhibitors of the aspartic protease endothiapepsin
Mondal M, Groothuis D, Hirsch A (2015)
DOI: 10.1039/C5MD00157A
Structure-Based Optimization of Inhibitors of the Aspartic Protease Endothiapepsin
Hartman A, Mondal M, Radeva N, Klebe G, Hirsch A (2015)
International journal of molecular sciences 16 (8): 19184-94DOI: 10.3390/ijms160819184
Supramolecular chemistry … and beyond
Hirsch A (2015)
Angewandte Chemie (International ed. in English) 54 (38): 11013-4DOI: 10.1002/anie.201506536
Validation of a homology model of Mycobacterium tuberculosis DXS: rationalization of observed activities of thiamine derivatives as potent inhibitors of two orthologues of DXS
Masini T, Lacy B, Monjas L, Hawksley D, Voogd A, Illarionov B, Iqbal A, Leeper F, Fischer M, Kontoyianni M, Hirsch A (2015)
Organic & biomolecular chemistry 13 (46): 11263-77DOI: 10.1039/c5ob01666e
Novel PqsR Inverse Agonists
Hamed M (2015)
Patent (EP 201523196)
2014
Development of inhibitors of the 2C-methyl-D-erythritol 4-phosphate (MEP) pathway enzymes as potential anti-infective agents
Masini T, Hirsch A (2014)
Journal of medicinal chemistry 57 (23): 9740-63DOI: 10.1021/jm5010978
Theoretical and structural analysis of long C-C bonds in the adducts of polycyanoethylene and anthracene derivatives and their connection to the reversibility of Diels-Alder reactions
Hirsch A, Reutenauer P, Le Moignan M, Ulrich S, Boul P, Harrowfield J, Jarowski P, Lehn J (2014)
Chemistry (Weinheim an der Bergstrasse, Germany) 20 (4): 1073-80DOI: 10.1002/chem.201303276
Structure-based design of inhibitors of the aspartic protease endothiapepsin by exploiting dynamic combinatorial chemistry
Mondal M, Radeva N, Köster H, Park A, Potamitis C, Zervou M, Klebe G, Hirsch A (2014)
Angewandte Chemie (International ed. in English) 53 (12): 3259-63DOI: 10.1002/anie.201309682
A natural-product switch for a dynamic protein interface
Scheepstra M, Nieto L, Hirsch A, Fuchs S, Leysen S, Lam C, het Panhuis L, van Boeckel C, Wienk H, Boelens R, …, Milroy L, Brunsveld L (2014)
Angewandte Chemie (International ed. in English) 53 (25): 6443-8DOI: 10.1002/anie.201403773
A doubly hermaphroditic chiral crown ether
Hirsch A, Sirlin C, Harrowfield J, Lehn J (2014)
DOI: 10.1039/C4CE00879K
De novo fragment-based design of inhibitors of DXS guided by spin-diffusion-based NMR spectroscopy
Masini T, Pilger J, Kroezen B, Illarionov B, Lottmann P, Fischer M, Griesinger C, Hirsch A (2014)
Chem. Sci. 5 (9): 3543-3551DOI: 10.1039/C4SC00588K
2013
Druggability of the enzymes of the non-mevalonate-pathway
Masini T, Kroezen B, Hirsch A (2013)
Drug Discovery Today 18 (23-24): 1256-62DOI: 10.1016/j.drudis.2013.07.003
Total synthesis, stereochemical elucidation and biological evaluation of Ac 2 SGL; a 1,3-methyl branched sulfoglycolipid from Mycobacterium tuberculosis
Geerdink D, Horst B, Lepore M, Mori L, Puzo G, Hirsch A, Gilleron M, Libero G, Minnaard A (2013)
DOI: 10.1039/C2SC21620E
Imidazole- and Benzimidazole-Based Inhibitors of the Kinase IspE: Targeting the Substrate-Binding Site and the Triphosphate-Binding Loop of the ATP Site
Mombelli P, Le Chapelain C, Munzinger N, Joliat E, Illarionov B, Schweizer W, Hirsch A, Fischer M, Bacher A, Diederich F (2013)
Exploiting specific interactions toward next-generation polymeric drug transporters
Wieczorek S, Krause E, Hackbarth S, Röder B, Hirsch A, Börner H (2013)
Journal of the American Chemical Society 135 (5): 1711-4DOI: 10.1021/ja311895z
2012
Total synthesis of (-)-doliculide, structure-activity relationship studies and its binding to F-actin
Matcha K, Madduri A, Roy S, Ziegler S, Waldmann H, Hirsch A, Minnaard A (2012)
ChemBioChem 13 (17): 2537-48DOI: 10.1002/cbic.201200512
The isoprenoid-precursor dependence of Plasmodium spp
van der Meer J, Hirsch A (2012)
Natural product reports 29 (7): 721-8DOI: 10.1039/c2np20013a
Metal-ion-induced shape switching: Stereoselective formation of a dinuclear Hg(II) double helicate from a hydrazonobis(acylhydrazone) ligand
Schaeffer G, Harrowfield J, Lehn J, Hirsch A (2012)
Biodynamers: self-organization-driven formation of doubly dynamic proteoids
Hirsch A, Buhler E, Lehn J (2012)
Journal of the American Chemical Society 134 (9): 4177-83DOI: 10.1021/ja2099134
Exploring the Ribose Sub-Pocket of the Substrate-Binding Site in Escherichia coli IspE: Structure-Based Design, Synthesis, and Biological Evaluation of Cytosines and Cytosine Analogues
Schütz A, Osawa S, Mathis J, Hirsch A, Bernet B, Illarionov B, Fischer M, Bacher A, Diederich F (2012)
2009
Bioconjugates to specifically render inhibitors water-soluble
Hirsch A, Diederich F, Antonietti M, Börner H (2009)
Soft Matter 6 (1): 88-91DOI: 10.1039/B915928B
2008
The Non-Mevalonate Pathway to Isoprenoid Biosynthesis: A Potential Source of New Drug Targets
Hirsch A, Diederich F (2008)
CHIMIA 62 (4): 226-230DOI: 10.2533/chimia.2008.226
Synthesis and characterization of cytidine derivatives that inhibit the kinase IspE of the non-mevalonate pathway for isoprenoid biosynthesis
Crane C, Hirsch A, Alphey M, Sgraja T, Lauw S, Illarionova V, Rohdich F, Eisenreich W, Hunter W, Bacher A, Diederich F (2008)
ChemMedChem 3 (1): 91-101DOI: 10.1002/cmdc.200700208
Inhibitors of the kinase IspE: structure-activity relationships and co-crystal structure analysis
Hirsch A, Alphey M, Lauw S, Seet M, Barandun L, Eisenreich W, Rohdich F, Hunter W, Bacher A, Diederich F (2008)
Organic & biomolecular chemistry 6 (15): 2719-30DOI: 10.1039/b804375b
2007
Phosphate recognition in structural biology
Hirsch A, Fischer F, Diederich F (2007)
Angewandte Chemie (International ed. in English) 46 (3): 338-52DOI: 10.1002/anie.200603420
Nonphosphate inhibitors of IspE protein, a kinase in the non-mevalonate pathway for isoprenoid biosynthesis and a potential target for antimalarial therapy
Hirsch A, Lauw S, Gersbach P, Schweizer W, Rohdich F, Eisenreich W, Bacher A, Diederich F (2007)
ChemMedChem 2 (6): 806-10DOI: 10.1002/cmdc.200700014
2006
Double conjugate addition of dithiols to propargylic carbonyl systems to generate protected 1,3-dicarbonyl compounds
Sneddon H, van den Heuvel A, Hirsch A, Booth R, Shaw D, Gaunt M, Ley S (2006)
The Journal of organic chemistry 71 (7): 2715-25DOI: 10.1021/jo052514s