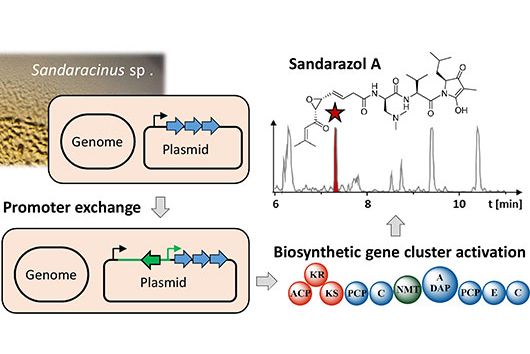
Saarbrücken, 11 July 2022 - One of the youngest research groups at HIPS is led by bioinformatician Dr Fabian Kern. His group is mainly concerned with the application of computer models in biomedicine and works under the name "Spatiotemporal Single-Cell Bioinformatics”. Together with the other groups at HIPS, Fabian wants to drive translational projects forward.
You recently started your own junior research group at HIPS. How was your start at HIPS?
All in all, great! I am thrilled by the many friendly and helpful people who work at HIPS and was therefore able to familiarise myself quickly, even if I needed a little help here and there in getting started. I would like to take this opportunity to thank everyone who helped me in such a pleasant way! And although I am not a trained pharmacist, I was able to find immediate scientific connections at the institute. As a bioinformatician with a doctorate in natural sciences from the Faculty of Medicine, I have been able to gain some experience in the field of biomedical applications and am now looking forward to the next stage, i.e. helping to shape the translational strategy at HIPS. It has always been important for me to tackle exciting new topics to constantly broaden my own horizon by getting to know other disciplines.
What are the main topics you will work on with your team? Why is this a useful extension to the activities already underway at HIPS and where do you see links?
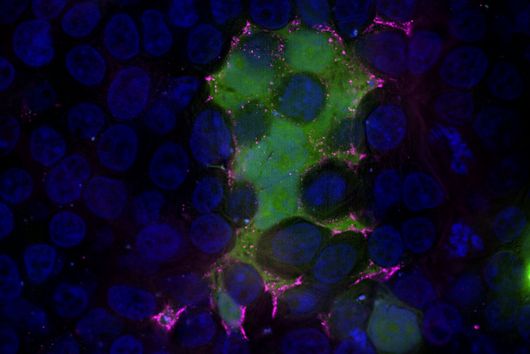
Well, I have had the good fortune to be able to support and evaluate many larger human sequencing projects, mainly in the field of transcriptomics, during my time at the group for clinical bioinformatics. Here I see potential to make our knowledge available to the established groups at HIPS and to provide advice on planning and implementation. In terms of content, we will dedicate ourselves to the analysis of single cell sequencing data, increasingly also with spatial representation (Spatial Transcriptomics). The really nice thing is that many of the methods in the laboratory as well as our computer models can easily be applied to the topics at HIPS. Together we can still learn a lot from bacteria, and the field of metatranscriptomics seems particularly promising to me.
Bioinformatics will grow significantly in the coming years as HIPS continues to expand - what opportunities and challenges do you see in this process?
It will indeed be an exciting challenge. I am pleased that some talented minds from bioinformatics have already found their way to HIPS. We are trained to work in an interdisciplinary way, which also means speaking many languages. On a typical day, for example, we meet with computer scientists in the morning and with natural scientists at lunchtime - this often brings a very diverse view to everyday problems. Now we have a great opportunity to combine the existing competences and to practice true innovation. I think this is also pretty much in line with Saarland's strategy for a rapid technological transformation. Especially at the beginning, however, it is important that we take the necessary time to get to know each other in order to understand better, where the individual research questions, requirements and skills lie. Moreover, I think we are all interested in finally crossing the famous "valley of death" of translational research.
Almost synchronously with the start of your own junior research group, you have now been awarded the Hans-and-Ruth-Giessen Prize. How do you plan to use the prize money of € 20,000? Does it open up opportunities for you that you wouldn't have had otherwise?
I am enormously pleased that the four-member board of the Hans-and-Ruth-Giessen-Foundation considered the project proposal for my research at HIPS to be worthy of funding and recommended it to the Foundation. As a young scientist after completing the PhD, one faces the challenging task of establishing oneself step by step as an independent researcher. And third-party funding is an essential part of this process. That's why this prize gives us a bit of freedom as a team to pursue our own bioinformatics research projects. First and foremost, we want to use infection models and spatial transcriptomics experiments to understand which cellular neighbourhoods are specific to pathogenic conditions. We see this as an excellent use case for neural networks that could help us find genetic signatures. At this point, we come back to the expertise at HIPS to synthesise new metabolites and drugs to modify cellular pathways. In terms of opening up opportunities, I will try to enhance our connections to other researchers and companies from the AI-Pharmacy field. Finally, the foundation cultivates an amazing alumni-network, many members of which share research experience in other countries and their contacts, as for example to the US.
What do you do when you are not working on your research?
I'm constantly eager exploring the surroundings and in Saarland there are countless opportunities to spend time in nature. This also helps me to sustain a balance toward my job. In my free time, I enjoy working with the local vegetable and herb gardens. In the evenings, I often read about current developments in astronomy - our universe is and remains a mystery that I can only marvel at time and time again.
Dr Alwin Hartman and Dr Yannic Nonnenmacher conducted the interview.