Want some more?
Researchers from the Department of Microbial Natural Products improve the production of a new drug candidate.
Saarbrücken, 31 August 2021 - New antibiotics are urgently needed in the fight against multi-resistant bacteria. The recently discovered darobactin is a potential candidate with resistance-breaking properties. This substance shows very good antimicrobial activity against some of the most problematic hospital germs. However, so far darobactin could only be produced in relatively small quantities, making detailed investigation and further development difficult. A team led by Dr Sebastian Groß from the Department of Microbial Natural Products has now addressed this problem. The scientists published their results in the journal Chemical Science.
Your new paper is about the natural product darobactin. Where does this molecule come from and why is it so interesting for you?
Darobactin is an antibiotic that was first described by Imai and colleagues in the journal Nature in 2019 and is naturally produced by the bacterium Photorhabdus khanii HGB1456. This bacterium lives in symbiosis with nematodes, i.e. threadworms, that attack and feed on insect larvae, among other things. The ability of P. khanii to produce darobactin gives the nematodes a decisive advantage, because the antibiotic kills harmful bacteria originating, for example, from the infested insect larvae. Their own intestinal bacteria, which are important for the nematodes, are immune to darobactin. Based on the analysis of genome data, it was predicted that darobactin could also be produced by a variety of other bacteria - including Yersinia pestis, the pathogen that causes the black plague.
Darobactin is interesting for us because, as an antibiotic, it acts specifically against Gram-negative bacteria. Gram-negative bacteria, in contrast to Gram-positive bacteria, have a double layer cell membrane, which is a difficult barrier for many antibiotics to overcome. It is therefore not surprising that many of the multidrug-resistant and difficult-to-treat hospital germs are Gram-negative bacteria. It is important for us to discover and develop new antibiotic structures in order to have effective treatment options for bacterial infections in the future. Darobactin has not only shown exceptionally potent antimicrobial activity against many Gram-negative bacteria that are potentially harmful to humans, but it also spares helpful bacteria that are found in the human gut and are important for digestion. Some of the antibiotics on the market today have the unpleasant side effect that they also attack the "good" bacteria in the intestine, whereas this problem might not occur with darobactin.
How can it be that darobactin only acts on Gram-negative bacteria?
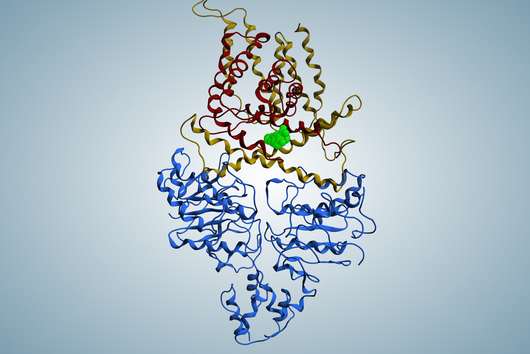
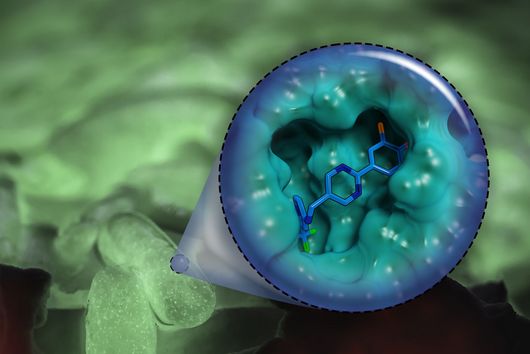
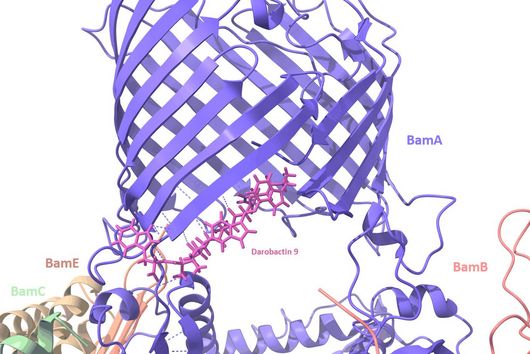
The specificity of darobactin against Gram-negative bacteria is based on its property to bind to a special protein located in the outer cell membrane of the bacterium and eventually inhibiting its function. Since only Gram-negative bacteria have this outer cell membrane, only these are combated by darobactin. The inhibited protein, BamA, is involved in building the outer cell membrane. If this function is lost, the bacteria are no longer able to survive. There is actually no other antibiotic used in human medicine that has BamA as its molecular target.
Why is it so important that the molecular target of darobactin is not yet exploited by any other antibiotic?
It has to do with the mechanism by which bacteria develop resistance to antibiotics. In the course of evolution, bacteria have developed various strategies to reduce the points of attack they offer antibiotics. For example, they can produce more cell membrane transporters to transport the antibiotic out of the cell so efficiently, that it can no longer reach its target in the cell. Another possibility would be the enzymatic modification of the antibiotic itself. This can happen either by cleaving the antibiotic, as in the case of penicillin, or by attaching functional groups, such as phosphate groups in the case of aminoglycoside antibiotics. This causes the respective antibiotic to lose its antimicrobial effect. Another type of resistance development is a change in the structure of the molecular target due to random mutations, so that the antibiotic simply can no longer bind to its target and thus loses its effect. Since there are several classes of antibiotics in human medicine that have the same molecular target in the bacterium, they may all lose their effectiveness at the same time. We call this phenomenon cross-resistance. However, this problem does not occur if the antibiotic targets a completely new structure, such as BamA in the case of darobactin.
In your study, you developed a strategy to enable the production of darobactin in larger quantities. Why is this necessary and how did you go about it?
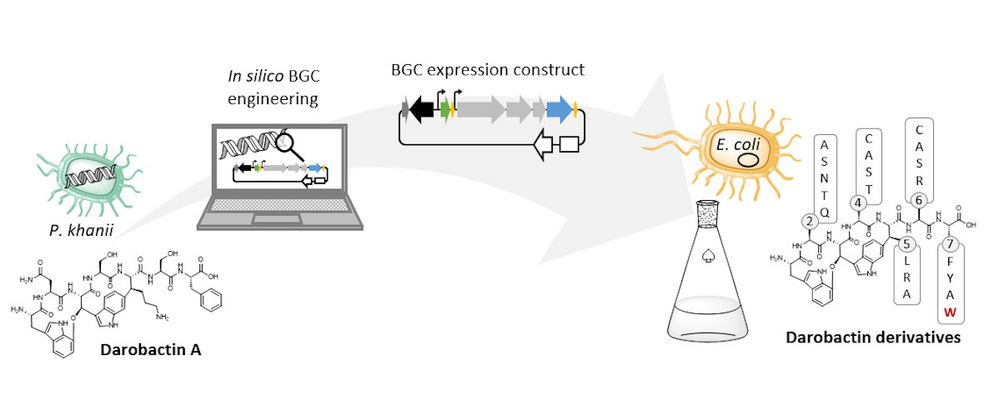
The natural producer of darobactin is a poorly characterised bacterium that produces very little darobactin under laboratory cultivation conditions. However, for a more precise characterisation of darobactin and the preclinical development of this substance class, larger amounts of substance are needed than could be provided by the existing processes with the natural producer. Since there are hardly any established protocols for rapid production improvement with the natural producer strain and the development of these methods would take a lot of time and effort, we decided to switch to Escherichia coli as an alternative producer. Although E. coli is one of the best-characterised microbes, it does not naturally produce darobactin. Therefore, we transferred all the genes that the natural producer needs to produce darobactin into E. coli. We virtually optimised the architecture of the biosynthetic gene cluster (BGC) to allow its expression in E. coli in order to be able to achieve the highest possible darobactin yield in the end. The BGC was then produced by DNA synthesis, cloned into an expression vector and introduced into E. coli. The resulting E. coli strain was indeed able to heterologously produce the derivative darobactin A, which was previously described by Imai and colleagues, but with a more than four-fold increased production rate compared to the natural producer.
When expressing the darobactin-BGC, you were able to discover 17 new derivatives in addition to the expected darobactin A. Was there anything exciting about this? Were you able to draw new conclusions from these results?
We did not discover the 17 new derivatives by chance, but deliberately aimed to produce them. We specifically altered a gene sequence in the darobactin-BGC to enable the production of these derivatives in E. coli. First, we roughly estimated the antimicrobial activity of the different derivatives against a small number of Gram-negative pathogens to see which derivatives had any antibacterial activity at all. Antibiotics often lose their activity upon structural modification. Among the darobactin derivatives we investigated, there were actually some promising candidates. Due to the complex darobactin purification process from the fermentation culture, we initially focused only on the derivative darobactin 9 and isolated it. Fortunately, compared to darobactin A, this derivative even showed improved antimicrobial activity against some important hospital pathogens, especially against various Pseudomonas aeruginosa and Acinetobacter baumannii strains, for which efficient treatment options do not exist anymore. While the activity profile of darobactin A against these strains still had gaps, we are already a significant step closer to the applicability in humans with darobactin 9. Nevertheless, a large number of experiments in the context of (pre-)clinical studies are still necessary until then.
Where do we go from here? What are the next steps?
We have set ourselves the goal of further improving the production yield. So far, we have only fermented our E. coli production strains in shake flasks. Cultivation under controlled and optimised conditions in a larger-scale fermenter still holds great potential for improvement.
In addition, we are planning, on the one hand, to further expand the structural diversity of the darobactins by producing more darobactin derivatives and, on the other hand, to purify and fully characterise some of the 17 already produced darobactin derivatives with promising antimicrobial activity.
Dr. Alwin Hartman and Dr. Yannic Nonnenmacher conducted the interview.
Original Paper
Sebastian Groß, Fabian Panter, Domen Pogorevc, Carsten E. Seyfert, Selina Deckarm, Chantal D. Bader, Jennifer Herrmann, Rolf Müller: Improved broad-spectrum antibiotics against Gram-negative pathogens via darobactin biosynthetic pathway engineering. Chem. Sci. 2021; doi: 10.1039/D1SC02725E