Saarbrücken, 10 March 2021 - Cystobactamids are a class of natural products with promising activity against multiresistant bacteria. Scientists at HIPS are therefore trying to optimize their properties so that they can be used to treat infections in humans. Together with his colleagues from the department of Prof. Rolf Müller, Dr. Sebastian Groß was able to elucidate the individual steps in cystobactamid biosynthesis, thus paving the way for further optimization of this class of molecules. Their results were now published in the journal Nature Communications.
Sebastian, your paper deals with the myxobacterial natural product class of cystobactamids. What makes this class of natural products interesting in the first place?
From a pharmaceutical point of view, cystobactamids are interesting because they are highly effective against Gram-negative bacteria. In contrast to Gram-positive bacteria, Gram-negative bacteria have an additional cell membrane surrounding the cell, which represents a barrier for many substances that is difficult to overcome. Since many multidrug-resistant and difficult-to-treat hospital pathogens are Gram-negative, the discovery and further development of novel antibiotics to combat these pathogens is important in order to ensure the availability of effective treatment options in the future.
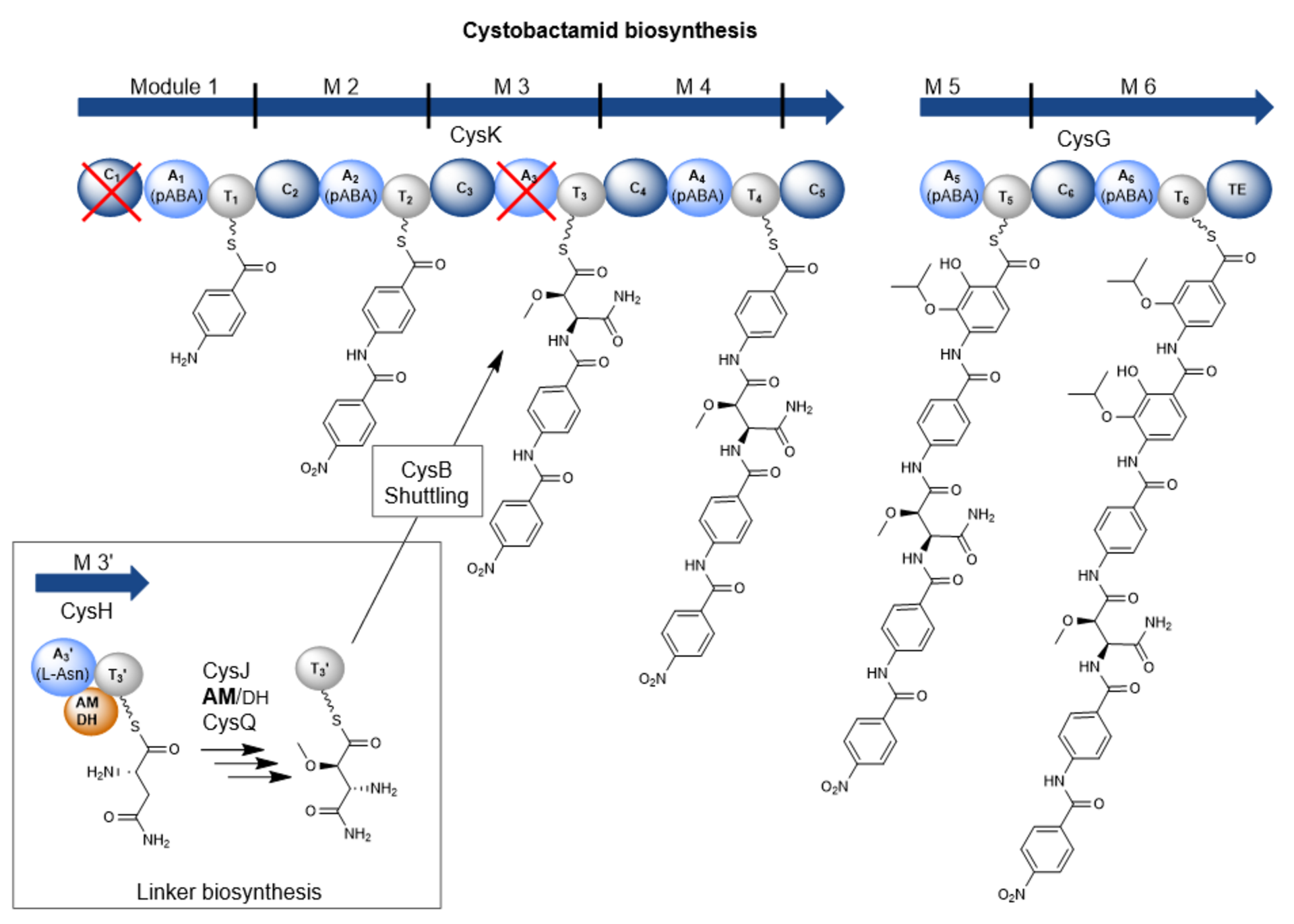
However, cystobactamids are also interesting molecules from a structural and biosynthetic perspective. They mainly consist of para-aminobenzoic acid (pABA) building blocks of which some are further modified. Those building blocks are rarely found in natural products. The pABA units are connected via a linker. In the known natural cystobactamid derivatives, different linker structures have been identified, such as the α-methoxy-L-isoasparagine unit, which is unique among all natural products known so far. Biosynthetically, cystobactamids are produced by a nonribosomal peptide synthetase (NRPS). However, previous publications suggest that the biosynthesis of cystobactamids harbors some differences compared to biosynthetic processes from already known NRPS systems. Therefore, we were interested in elucidating the individual steps of cystobactamid biosynthesis, especially regarding the unique α-methoxy-L-isoasparagine linker.
How did you go about elucidating each step?
In a first approach, we produced the enzymes involved in the biosynthesis of cystobactamids individually in Escherichia coli, purified them, and then examined their activity in the test tube (in vitro). Depending on the availability of substrates, cofactors, and other reaction conditions, we were able to artificially reconstitute some biosynthetic reactions. In a complementary set of experiments, we removed certain genes or parts of genes encoding enzymes involved in biosynthesis from the biosynthetic gene cluster (BGC). This resulted in changes in the cystobactamid production profile of the bacterium, which allowed us to draw conclusions about the missing or altered biochemical reaction steps.
In the paper you use bacteria of the species Myxococcus xanthus for the production of cystobactamids and for the elucidation of their biosynthesis. However, M. xanthus is not a native producer of cystobactamids. How does this work and why did you choose this approach?
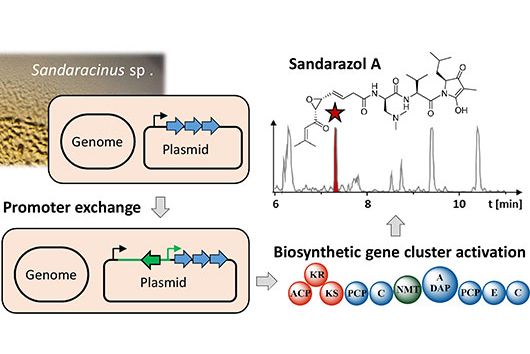
That's correct. There are various strains of myxobacteria that are capable of producing cystobactamids. These include, for example, strains of the genera Cystobacter, Myxococcus and Corallococcus. However, all of these strains are only capable to produce cystobactamids at relatively low rates under laboratory conditions. Therefore, we switched to the very well characterized myxobacterial model strain M. xanthus DK1622 for our experiments. This strain does not naturally produce cystobactamids – therefore, we first had to transfer the entire BGC, containing all genes required for cystobactamid production, into it.
For this purpose, we computationally (in silico) designed a modified BGC based on the DNA sequence of the native cystobactamid producer Cystobacter velatus Cbv34. The individual fragments of this modified BGC were then chemically synthesized and assembled in the laboratory before we introduced it into M. xanthus DK1622. In the end, cystobactamid production rates in this strain were even significantly higher than previously published values from the native strains.
You describe the elucidation of the individual steps in the biosynthesis of cystobactamids. Why is it important to know these steps in such detail?
Understanding the biosynthetic steps allows us to manipulate the cystobactamid biosynthesis in a targeted way, for example, in order to produce new derivatives with improved properties or to identify and eliminate potential rate-limiting biosynthetic steps.
In addition, the elucidation of cystobactamid biosynthesis helps to improve our general understanding of NRPS systems. The prototypical functioning and classification of NRPS systems described in the literature is based on the study of only a few systems, e.g. from actinobacteria. However, myxobacteria are only distantly related to actinobacteria and the functioning of myxobacterial NRPS systems seems to be significantly different from other NRPS systems. This partially results from the unique structures of natural products found only in myxobacteria. Our results help to elucidate these different and novel functionalities of myxobacterial NRPS systems.
What are the special features of cystobactamid biosynthesis? Are there any special or unique features?
During early NRPS research, attempts were made to establish rules for their functioning, which are generally applicable. For example, the colinearity rule states that each functional subunit (so-called module) of an NRPS is responsible for the incorporation of a single building block into the final product. In addition, the processivity rule states that biosynthesis starts with the first module and sequentially continues to the terminal module. Over time, however, more and more exceptions to these rules have been found, as is now the case in cystobactamid biosynthesis.
The major peculiarity here is the biosynthesis of the linker unit. This is carried out by an independent NRPS module that is not part of the two major NRPS proteins. This module acts in conjunction with two other, external enzymes and thus contributes to the structural diversity in the linker group of cystobactamids. In addition, a bifunctional domain frequently found in natural product biosyntheses, but never experimentally characterized before, has been described in this NRPS module. This domain can catalyze either the dehydration or the isomerization of L-asparagine. Which reaction is favored depends on the activity of an oxygenase enzyme that hydroxylates L-asparagine and consequently prevents dehydration to β-cyano-L-alanine. Another distinctive feature is the transport process of the linker unit from the independent NRPS module to the NRPS itself, which is carried out via a special transport protein. These biosynthetic processes are in direct contradiction with the processivity rule.
Furthermore, even after artificial removal of the linker transport protein, we detected the production of a cystobactamid tripeptide produced by the three terminal modules of the NRPS. This means that this tripeptide is neither a degradation product from full-length cystobactamids, nor does the biosynthesis start from the first module, but it likely starts from a module within the NRPS. Thus, cystobactamid biosynthesis also "violates" the colinearity rule.
Is the biosynthesis of cystobactamids now fully elucidated or are there still more surprises to be expected in future experiments?
There are still one or two open questions that we have not been able to answer with our experiments so far. For example, we have shown the function of the bifunctional NRPS domain involved in cystobactamid linker biosynthesis only via its deletion. Deletion, in turn, can lead to the loss of essential protein-protein interactions and thus affect the activity of other enzymes that function only in complex. In future experiments, we aim to exchange specific catalytically active amino acids in this domain by selectively introducing point mutations in the BGC. Ideally, we would inactivate one of the two functions without affecting other enzymes that require the intact structure of the bifunctional domain.
Where do we go from here? What are the next steps in brining cystobactamid closer to clinical application?
We will try to apply the knowledge we have gained about cystobactamid biosynthesis in a way that directs the cystobactamid production profile in M. xanthus DK1622 towards more active derivatives. So far, we have been partially successful in doing this by removing the bifunctional NRPS domain. However, we suspect that the linker biosynthesis and transport of the linker to the NRPS are rate-limiting reactions. One planned approach is to reactivate an inactive NRPS module whose function has been taken over by the independent NRPS module. By doing so, we hope to improve cystobactamid production so that our system can be used as an alternative to the established chemical synthesis of cystobactamids.
Regarding the development of cystobactamid into a marketable drug, several new derivatives that are more active than natural cystobactamids have already been produced using medicinal chemistry. These promising derivatives are currently being tested for their clinical applicability.
Dr. Yannic Nonnenmacher conducted the interview.
Original Publication:
Sebastian Groß, Bastien Schnell, Patrick A. Haack, David Auerbach, and Rolf Müller: In vivo and in vitro reconstitution of unique key steps in cystobactamid antibiotic biosynthesis. Nature Communications, 2021. doi: 10.1038/s41467-021-21848-3